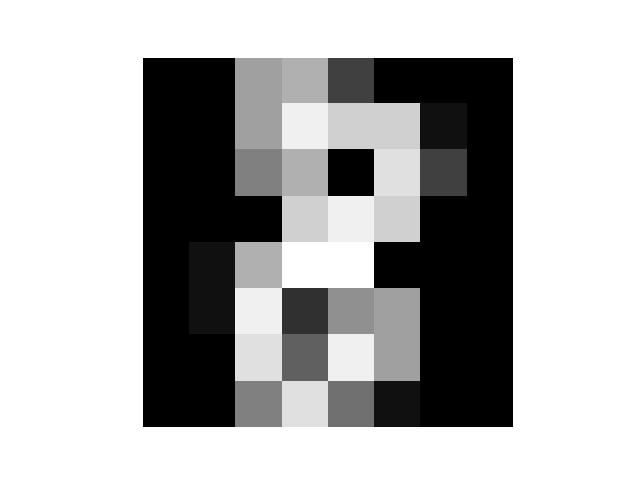Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
Naive Bayes classifier - Visualisation¶
Naive Bayes classifier
Computing the posterior probability of x being from class c using Bayes rule.
This script demonstrates Naive Bayes classifier using three examples
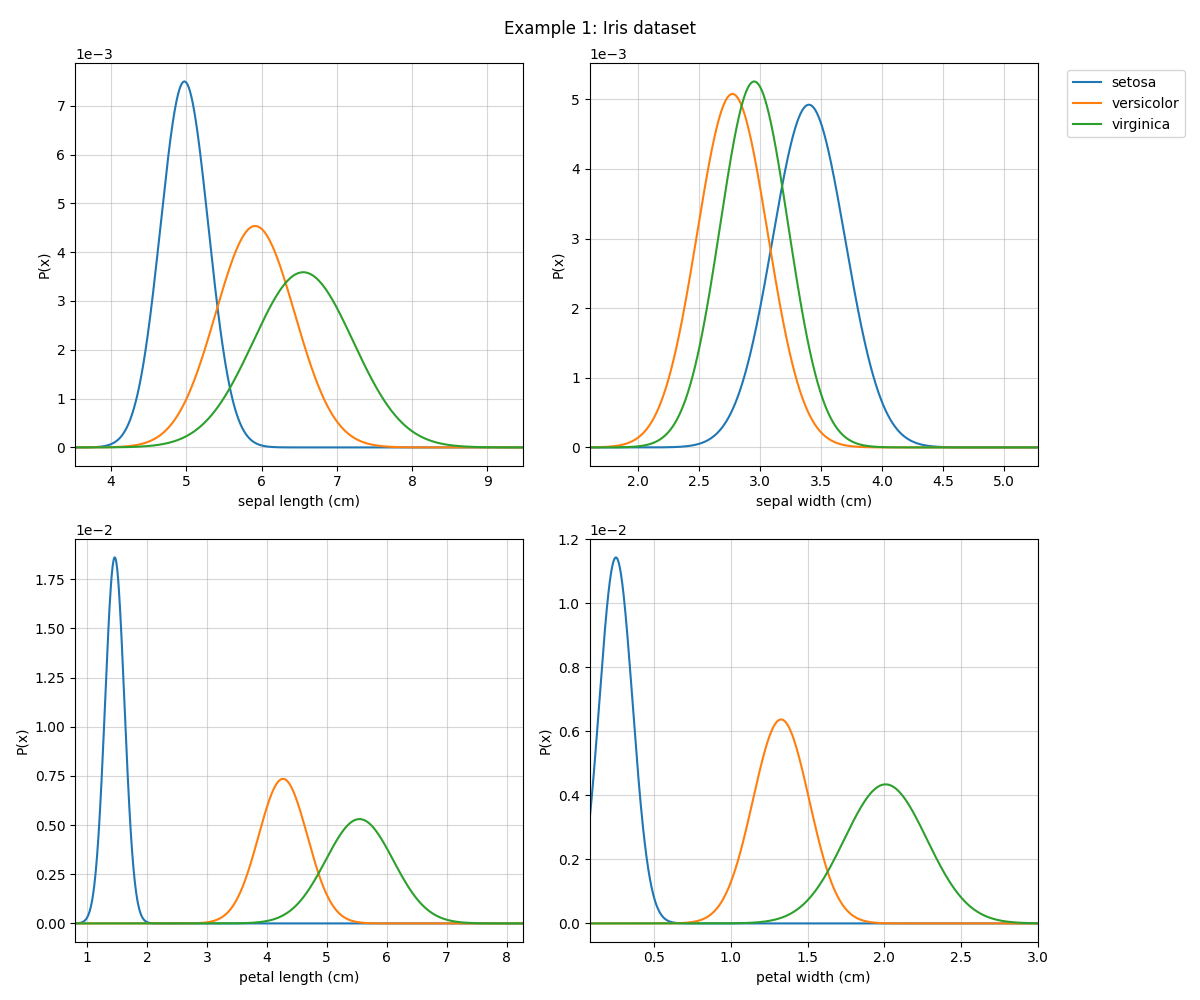
Example 1: Iris dataset
Example 2: Breast Cancer
Example 3: Digit Classification
# libraries
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.model_selection import train_test_split
import spkit
print('spkit version :', spkit.__version__)
from spkit.ml import NaiveBayes
spkit version : 0.0.9.7
Example 1 : Iris dataset¶
data = datasets.load_iris()
X = data.data
y = data.target
Xt,Xs,yt,ys = train_test_split(X,y,test_size=0.3)
print('Shapes: ',Xt.shape,yt.shape,Xs.shape,ys.shape)
# Fitting model (Estimating the parameters)
model = NaiveBayes()
model.fit(Xt,yt)
# ## Prediction and Accuracy
ytp = model.predict(Xt)
ysp = model.predict(Xs)
print('Training Accuracy : ',np.mean(ytp==yt))
print('Testing Accuracy : ',np.mean(ysp==ys))
# Predicting probabilities
ytpr = model.predict_prob(Xt)
yspr = model.predict_prob(Xs)
print(ytpr[0])
print(model.predict(Xs[0]), model.predict_prob(Xs[0]))
# Parameters :: $\mu$, $\sigma$
print('model parameters')
print(model.parameters)
# Visualizing the distribution
# Setting the names of classes and features (Optional)
model.set_class_labels(data['target_names'])
model.set_feature_names(data['feature_names'])
fig = plt.figure(figsize=(12,10))
model.VizPx(show=False)
plt.suptitle('Example 1: Iris dataset')
plt.tight_layout()
plt.show()
Shapes: (105, 4) (105,) (45, 4) (45,)
Training Accuracy : 0.9619047619047619
Testing Accuracy : 0.9333333333333333
[6.40553556e-10 1.17910306e-04 9.99882089e-01]
[0] [[9.99999989e-01 5.43110935e-09 5.43110703e-09]]
model parameters
{0: {'mu': array([4.97428571, 3.4 , 1.46285714, 0.25142857]), 'sig': array([0.10076735, 0.08914286, 0.02576327, 0.01164082]), 'prior': 0.3333333333333333}, 1: {'mu': array([5.91515152, 2.77272727, 4.26969697, 1.32727273]), 'sig': array([0.27522498, 0.08380165, 0.16514233, 0.03349862]), 'prior': 0.3142857142857143}, 2: {'mu': array([6.55405405, 2.95135135, 5.54594595, 2.00810811]), 'sig': array([0.43978086, 0.07817385, 0.31707816, 0.07209642]), 'prior': 0.3523809523809524}}
Example 2: Breast Cancer¶
data = datasets.load_breast_cancer()
X = data.data
y = data.target
Xt,Xs,yt,ys = train_test_split(X,y,test_size=0.3)
print(Xt.shape,yt.shape,Xs.shape,ys.shape)
# ## Fitting model (estimating the parameters)
model = NaiveBayes()
model.fit(Xt,yt)
# ## Accuracy
ytp = model.predict(Xt)
ysp = model.predict(Xs)
print('Training Accuracy : ',np.mean(ytp==yt))
print('Testing Accuracy : ',np.mean(ysp==ys))
# Parameters :: $\mu$, $\sigma$
print('model parameters')
print(model.parameters[0])
model.set_class_labels(data['target_names'])
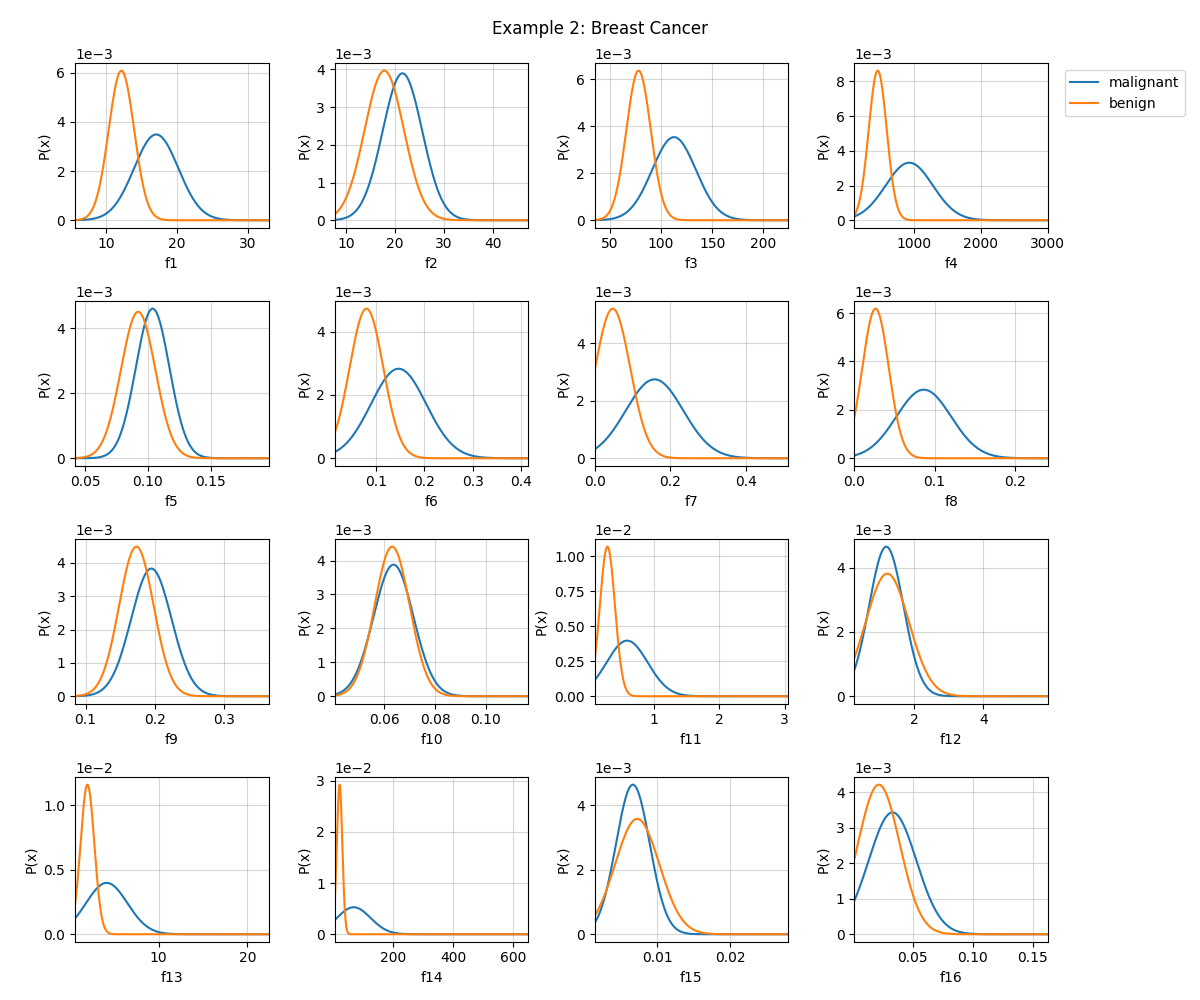
# Visualizing first 16 features
fig = plt.figure(figsize=(12,10))
model.VizPx(nfeatures=range(16),show=False)
plt.suptitle('Example 2: Breast Cancer')
plt.tight_layout()
plt.show()
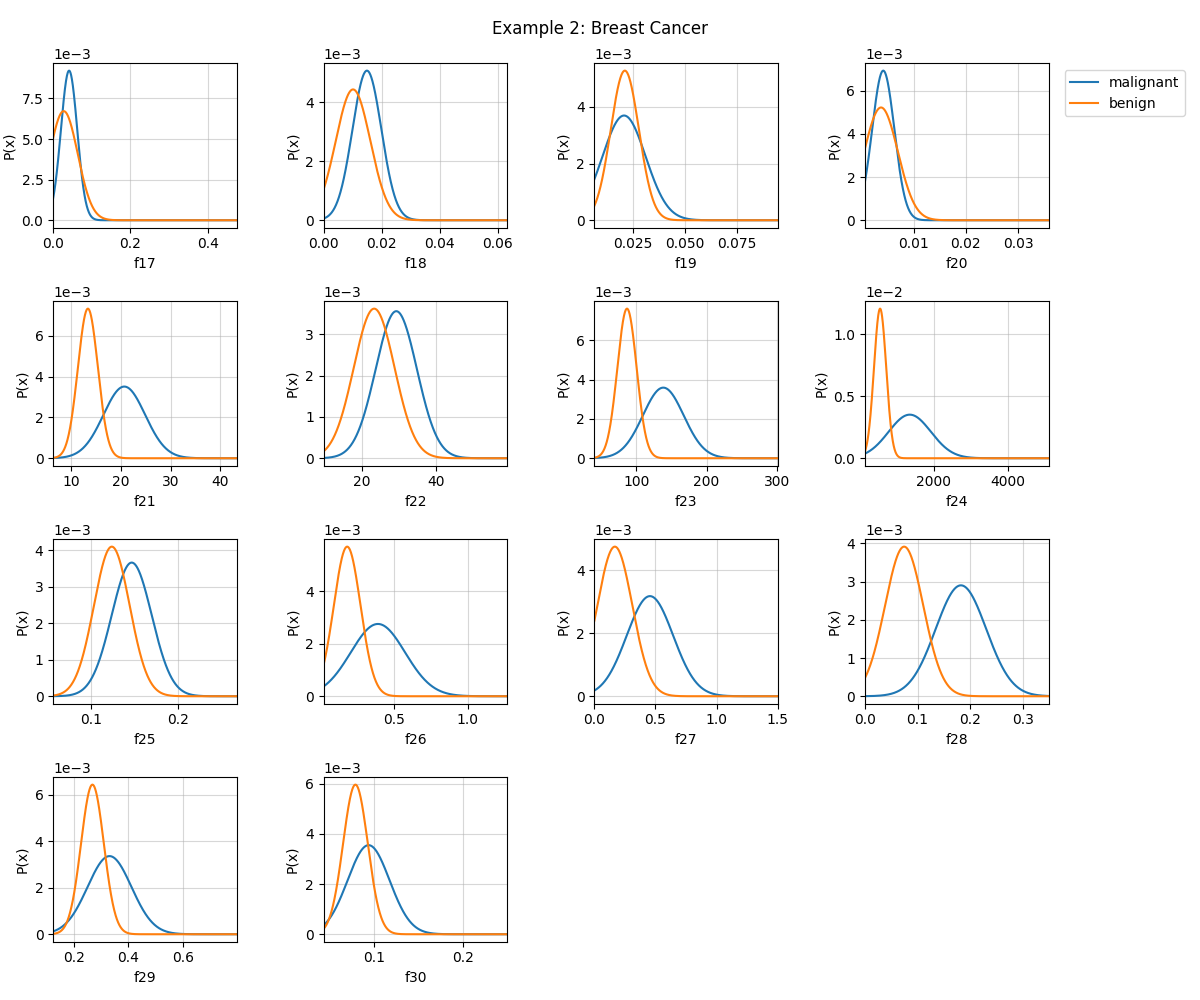
# Visualizing next 14 features
fig = plt.figure(figsize=(12,10))
model.VizPx(nfeatures=range(16,30),show=False)
plt.suptitle('Example 2: Breast Cancer')
plt.tight_layout()
plt.show()
(398, 30) (398,) (171, 30) (171,)
Training Accuracy : 0.9321608040201005
Testing Accuracy : 0.9415204678362573
model parameters
{'mu': array([1.70467143e+01, 2.15247857e+01, 1.12635429e+02, 9.31802143e+02,
1.03847500e-01, 1.46936357e-01, 1.58429357e-01, 8.64501429e-02,
1.95218571e-01, 6.35545000e-02, 5.87085714e-01, 1.20458429e+00,
4.16767143e+00, 6.83693571e+01, 6.62769286e-03, 3.34023214e-02,
4.12837857e-02, 1.48983929e-02, 2.05943500e-02, 4.20243571e-03,
2.06528571e+01, 2.93108571e+01, 1.38365786e+02, 1.35655286e+03,
1.46919214e-01, 3.90022929e-01, 4.54795571e-01, 1.82548643e-01,
3.31425714e-01, 9.40867857e-02]), 'sig': array([9.76761349e+00, 1.62777392e+01, 4.57830703e+02, 1.23242710e+05,
1.79038660e-04, 3.23655220e-03, 5.77591931e-03, 1.17075703e-03,
8.54116184e-04, 6.22985933e-05, 9.97596042e-02, 2.43670494e-01,
5.40617123e+00, 3.14229457e+03, 5.36246133e-06, 3.91651263e-04,
4.46363544e-04, 2.49955426e-05, 1.09162800e-04, 4.52310239e-06,
1.75820447e+01, 3.13523207e+01, 8.43107083e+02, 3.26871981e+05,
5.24808469e-04, 3.41448475e-02, 3.61006737e-02, 2.31270669e-03,
6.43498830e-03, 5.48420363e-04]), 'prior': 0.35175879396984927}
Example 3:: Digit Classification¶
data = datasets.load_digits()
X = data.data
y = data.target
# Avoiding features with zero variance (contant value)
# X = X[:,X.var(0)>0]
Xt,Xs,yt,ys = train_test_split(X,y,test_size=0.3)
print(Xt.shape,yt.shape,Xs.shape,ys.shape)
# Fitting model (estimating the parameters)
model = NaiveBayes()
model.fit(Xt,yt)
# Accuracy
ytp = model.predict(Xt)
ysp = model.predict(Xs)
print('Training Accuracy : ',np.mean(ytp==yt))
print('Testing Accuracy : ',np.mean(ysp==ys))
# Predicting probablities
print(model.predict(Xs[0]), model.predict_prob(Xs[0]))
plt.imshow(Xs[0].reshape([8,8]),cmap='gray')
plt.axis('off')
plt.show()
print('Prediction',model.predict(Xs[0]))
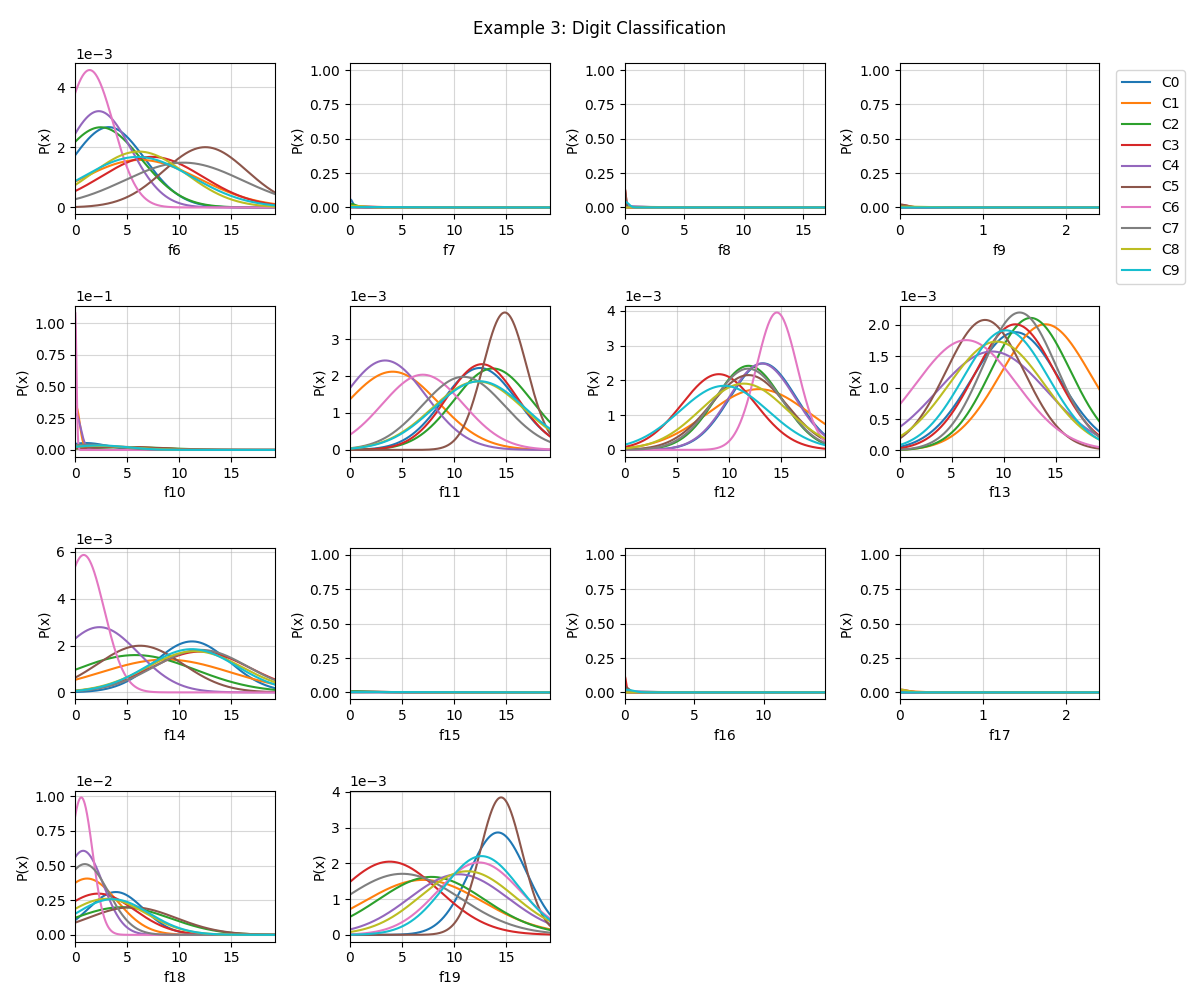
# Visualizing
fig = plt.figure(figsize=(12,10))
model.VizPx(nfeatures=range(5,19),show=False)
plt.suptitle('Example 3: Digit Classification')
plt.tight_layout()
plt.show()
(1257, 64) (1257,) (540, 64) (540,)
Training Accuracy : 0.7955449482895783
Testing Accuracy : 0.75
[8] [[5.20647505e-20 4.40255105e-12 3.70948008e-14 5.20647506e-20
5.20647505e-20 5.20647505e-20 5.20647505e-20 5.20647505e-20
1.00000000e+00 5.20647505e-20]]
Prediction [8]
Total running time of the script: (0 minutes 8.422 seconds)
Related examples
Decision Trees with shrinking capability - Regression example
Decision Trees with visualisations while buiding tree
Decision Trees with shrinking capability - Classification example
Decision Trees without converting Catogorical Features