spkit.mea.activation_repol_time_loc¶
- spkit.mea.activation_repol_time_loc(X, fs=25000, at_range=[None, None], rt_range=[0.5, None], method='min_dvdt', gradient_method='fdiff', sg_window=11, sg_polyorder=3, gauss_window=0, gauss_itr=1, plot=False, plot_dur=2, **kwargs)¶
Computing Activation and Repolarisation Time together
Computing Activation Time and Repolarisation Time of multi-channel signals
Activation Time in cardiac electrograms refered to as time at which depolarisation of cells/tissues/heart occures. In contrast to ‘Activation Time’ in cardiac electrograms, Repolarisation Time, also refered as Recovery Time, indicates a time at which repolarisation of cells/tissues/heart occures.
For biological signals (e.g. cardiac electorgram), an activation time in signal is reflected by maximum negative deflection, which is equal to min-dvdt, if signal is a volatge signal and function of time x = v(t) and repolarisation time is again a reflected by maximum deflection (mostly negative), after activation occures.
However, simply computing derivative of signal is sometime misleads the activation and reoolarisation time location, due to noise, so derivative of a given signal has be computed after pre-processing. Repolarisation Time is often very hard to detect reliably, due to very small electrogram, which is mostly lost in noise.
- Parameters:
- Xnd-array
Single Cycle of each channel containing EGM
with shape = (nch,n),
where nch: number of channels, n: number of samples,
- fs: int, default=25000
sampling frequency of signal,
- t_range: list of [t0 (ms),t1 (ms)]
range of time to restrict the search of activation time during t0 ms to t1 ms
if
t_range=[None,None], whole input signal is considered for searchif
t_range=[t0,None], excluding signal before t0 msif
t_range=[None,t1], excluding signal after t1 ms for search
- rt_range: list of [t0 (ms),t1 (ms)]
range of time to restrict the search of repolarisation time
check
spkit.get_activation_timeandspkit.get_repolarisation_timefor more details
- method: str, default=”min_dvdt”
Method to compute activation time
one of (“max_dvdt”, “min_dvdt”, “max_abs_dvdt”)
for more detail
spkit.get_activation_time
- gradient_method: str, default=’fdiff’
Method to compute gradient of signal
one of (“fdiff”, “fgrad”, “npdiff”,”sgdiff”,”sgdrift_diff”,”sgsmooth_diff”, “gauss_diff”)
check
spkit.signal_diff
Note
- Same ‘method’ and ‘gradient_method’ is applied to both AT and RT computation.
To use different methods use
spkit.get_activation_timeandspkit.get_repolarisation_timeseperatly
- Parameters for gradient_method:
used if gradient_method in one of (“sgdiff”,”sgdrift_diff”,”sgsmooth_diff”, “gauss_diff”)
sg_window: sgolay-filter’s window length
sg_polyorder: sgolay-filter’s polynomial order
gauss_window: window size of gaussian kernel for smoothing,
check help(signal_diff) from sp.signal_diff
- plot:int, default=False
If true, plot 3 figures for each channel
Full signal trace with activation time and repolarisation time
Segment of signal from loc-plot_dur to loc+plot_dur for activation time
Segment of signal from loc-plot_dur to loc+plot_dur for repolarisation time
Derivative of signal, with with activation time and repolarisation time
- plot_dur: scalar, default=2,
duration in seconds to plot, if plot=True
segment of signal from loc-plot_dur to loc+plot_dur
default=2s
- gradient_method: Method to compute gradient of signal
one of (“fdiff”, “fgrad”, “npdiff”,”sgdiff”,”sgdrift_diff”,”sgsmooth_diff”, “gauss_diff”) check help(signal_diff) from sp.signal_diff
- NOTE: Same ‘method’ and ‘gradient_method’ is applied to both AT and RT computation.
To use different methods use ‘get_activation_time’ and ‘get_repolarisation_time’ seperatly
- Parameters for gradient_method:
- used if gradient_method in one of (“sgdiff”,”sgdrift_diff”,”sgsmooth_diff”, “gauss_diff”)
sg_window: sgolay-filter’s window length sg_polyorder: sgolay-filter’s polynomial order gauss_window: window size of gaussian kernel for smoothing, check help(signal_diff) from sp.signal_diff
- Returns:
- at_loc: 1d-array
array of length=nch, location of activation time as index
to convert it in seconds, at_loc/fs
- rt_loc: 1d-array
array of length=nch, location of repolarisation time as index
to convert it in seconds, rt_loc/fs
Examples
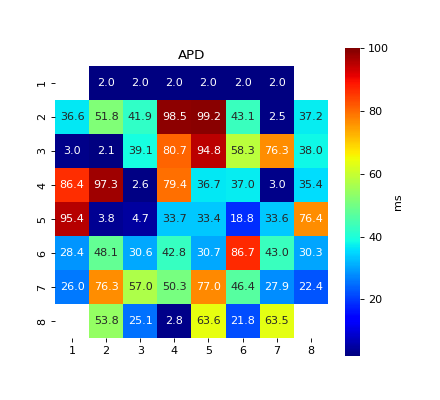
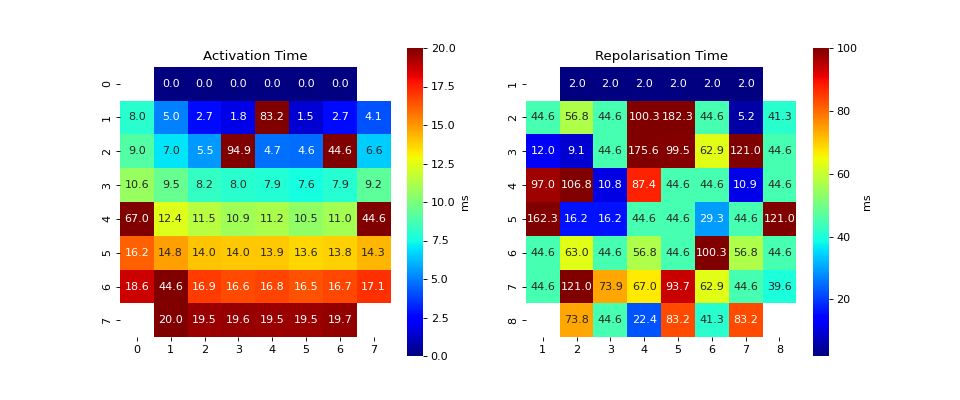
#sp.mea.activation_repol_time_loc import numpy as np import matplotlib.pyplot as plt import os, requests import spkit as sp # Download Sample file if not done already file_name= 'MEA_Sample_North_1000mV_1Hz.h5' if not(os.path.exists(file_name)): path = 'https://spkit.github.io/data_samples/files/MEA_Sample_North_1000mV_1Hz.h5' req = requests.get(path) with open(file_name, 'wb') as f: f.write(req.content) ############################## # Step 1: Read File fs = 25000 X,fs,ch_labels = sp.io.read_hdf(file_name,fs=fs,verbose=1) ############################## # Step 2: Stim Localisation stim_fhz = 1 stim_loc,_ = sp.mea.get_stim_loc(X,fs=fs,fhz=stim_fhz, plot=0,verbose=0,N=None) ############################## # Step 3: Align Cycles exclude_first_dur=2 dur_after_spike=500 exclude_last_cycle=True XB = sp.mea.align_cycles(X,stim_loc,fs=fs, exclude_first_dur=exclude_first_dur,dur_after_spike=dur_after_spike, exclude_last_cycle=exclude_last_cycle,pad=np.nan,verbose=True) print('Number of EGMs/Cycles per channel =',XB.shape[2]) ############################## # Step 4: Average Cycles egm_number = -1 if egm_number<0: X1B = np.nanmean(XB,axis=2) print(' -- Averaged All EGM') else: # egm_number should be between from 0 to 'Number of EGMs/Cycles per channel ' assert egm_number in list(range(XB.shape[2])) X1B = XB[:,:,egm_number] print(' -- Selected EGM ->',egm_number) print('EGM Shape : ',X1B.shape) ############################## # Step 5-6: Activation and Repolarisation Time at_range = [0,100] rt_range = [2,100] at_loc, rt_loc = sp.mea.activation_repol_time_loc(X1B,fs=fs,at_range=at_range, rt_range=rt_range) at_loc_ms = 1000*at_loc/fs rt_loc_ms = 1000*rt_loc/fs print(at_loc_ms) print(rt_loc_ms) ############################## # Step 7: APD apd_ms = rt_loc_ms-at_loc_ms AT_grid = sp.mea.arrange_mea_grid(at_loc_ms, ch_labels=ch_labels) RT_grid = sp.mea.arrange_mea_grid(rt_loc_ms, ch_labels=ch_labels) APD_grid = sp.mea.arrange_mea_grid(apd_ms, ch_labels=ch_labels) fig, ax = plt.subplots(1,2, figsize=(12,5)) sp.mea.mat_1_show(AT_grid,vmax=20, label = ('ms'),ax=ax[0]) ax[0].set_title('Activation Time') sp.mea.mat_1_show(RT_grid,vmax=100,label = ('ms'),ax=ax[1]) ax[1].set_title('Repolarisation Time') plt.show()
fig, ax = plt.subplots(figsize=(5.5,5)) sp.mea.mat_1_show(APD_grid,vmax=100, label = ('ms'),ax=ax) ax.set_title('APD') plt.show()