spkit.get_activation_time¶
- spkit.get_activation_time(x, fs=1, method='min_dvdt', gradient_method='fdiff', sg_window=11, sg_polyorder=3, gauss_window=0, gauss_itr=1, **kwargs)¶
Get Activation Time based on Gradient
Get Activation Time based on Gradient
Activation Time in cardiac electrograms refered to as time at which depolarisation of cells/tissues/heart occures.
For biological signals (e.g. cardiac electorgram), an activation time in signal is reflected by maximum negative deflection, which is equal to min-dvdt, if signal is a volatge signal and function of time x = v(t) However, simply computing derivative of signal is sometime misleads the activation time locatio, due to noise, so derivative of a given signal has be computed after pre-processing
- Parameters:
- x: 1d-array,
input signal
- fs: int,
sampling frequency, default fs=1, in case only interested in loc
- methoddefault = “min_dvdt”
Method to compute Activation Time, “min_dvdt” is the used in literature, however, it depends on the kind of signal. For Action Potential Signal, “max_dvdt” is considered as Activation Time for Unipolar Electrogram, “min_dvdt” is consodered. it can be chosen one of {“max_dvdt”, “min_dvdt”, “max_abs_dvdt”},
Some literation suggests to use max_dvdt instead of min_dvdt, but mostly agree on min_dvdt
- gradient_method: default =”fdiff”,
{“fdiff”, “fgrad”,”sgdiff”,”sgdrift_diff”,”sgsmooth_diff”, “gauss_diff”}
Method to compute gradient of signal
one of {“fdiff”, “fgrad”, “npdiff”,”sgdiff”,”sgdrift_diff”,”sgsmooth_diff”, “gauss_diff”}
check
signal_difffor more details on the methodif signal is noisy try “sgsmooth_diff” or “gauss_diff”
- Parameters for gradient_method:
used if gradient_method in one of {“sgdiff”,”sgdrift_diff”,”sgsmooth_diff”, “gauss_diff”}
sg_window: sgolay-filter’s window length
sg_polyorder: sgolay-filter’s polynomial order
gauss_window: window size of gaussian kernel for smoothing,
check
signal_difffor more details on the method
- Returns:
- atactivation time (ms)
- loc: index
- mag: magnitude of deflection at loc
- dxderivative of signal x
Notes
#TODO
References
Examples
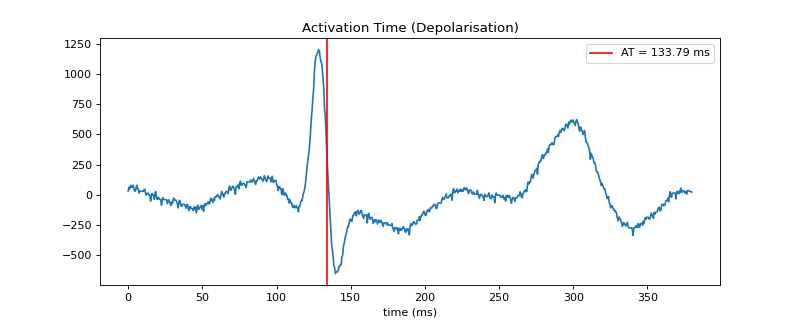
#sp.get_activation_time import numpy as np import matplotlib.pyplot as plt import spkit as sp x, fs = sp.data.ecg_sample(sample=1) x = sp.filterDC_sGolay(x,window_length=fs//2+1) #x = sp.filter_smooth_gauss(x,window_length=31) x = x[int(0.02*fs):int(0.4*fs)] t = 1000*np.arange(len(x))/fs # It is a good idea to smooth the signal or use gradient_method = gauss_diff at,loc,mag,dx= sp.get_activation_time(x,fs=fs,method='min_dvdt',gradient_method='fdiff') plt.figure(figsize=(10,4)) plt.plot(t,x) plt.axvline(at,color='r',label=f'AT = {at:0.2f} ms') plt.legend() plt.xlabel('time (ms)') plt.title('Activation Time (Depolarisation)') plt.show()