5.1. EEG: Electroencephalography¶
5.1.1. Preprocessing¶
5.1.1.1. Filtering¶
5.1.1.2. Power computation¶
5.1.1.3. Rhythmic Features¶
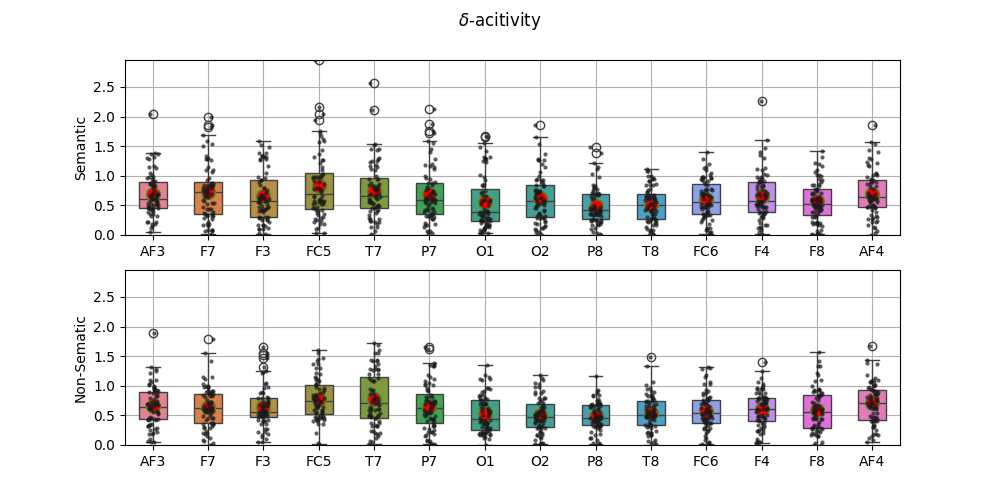
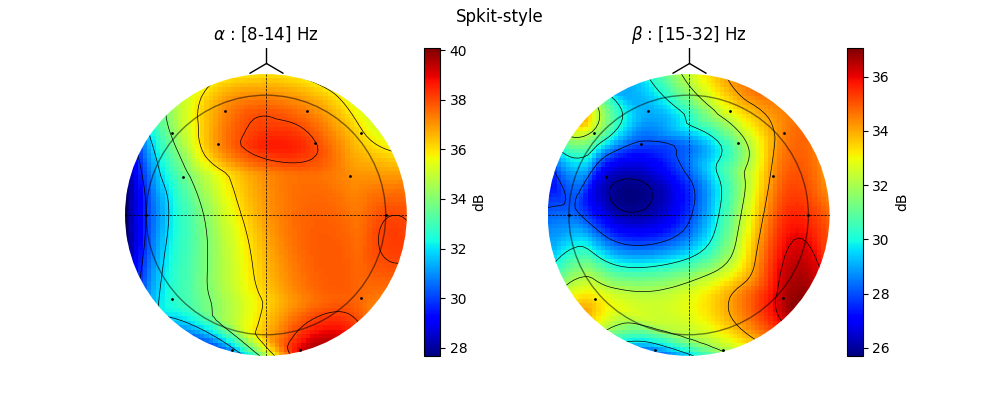
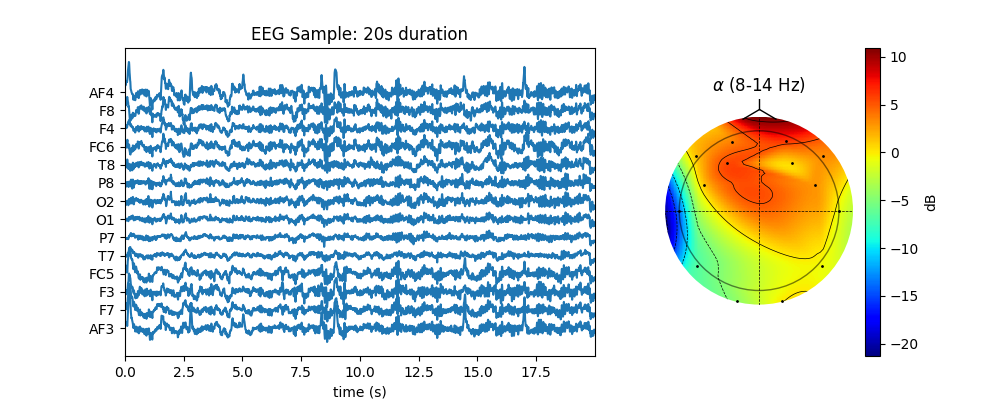
5.1.1.4. Topographic Map¶
5.1.2. Working with different files/frameworks¶
Several file formats are used for storing EEG Data. Some are specifically designed to support device recording systems, and some are there to support specific frameworks and tools, such as eeglab/mne, etc. Even though these file formats/tools/frameworks are efficient for a particular field/scope, they limit the researchers/users’ ability to explore data freely (at least that’s how I feel). And sometimes, they even limit the understanding of the data itself.
We prefer to extract raw signals from these files to process. This approach gives researchers/users much more independence, as after extracting signals as arrays (numpy.arrays), one can apply any signal processing algorithm, from a wide-variety of fields or even to custom-built algorithms.
5.1.2.1. Read from EDF Files¶
Spkit have function spkit.io.read_bdf that supports reading EDF/BDF files of EEG.
Here is an example
5.1.2.2. Work with MNE Raw Object¶
MNE is one of the tool used for EEG data processing. It allows to read several file formates as Raw Object, which further includes many analysis algorithms
Here is an example of reading MNE Raw object, extract EEG data from it, process it with ATAR algorithm and put EEG data back to MNE Raw Object for further processing and analysis.
Raw EEG Data |
After Applying ATAR |
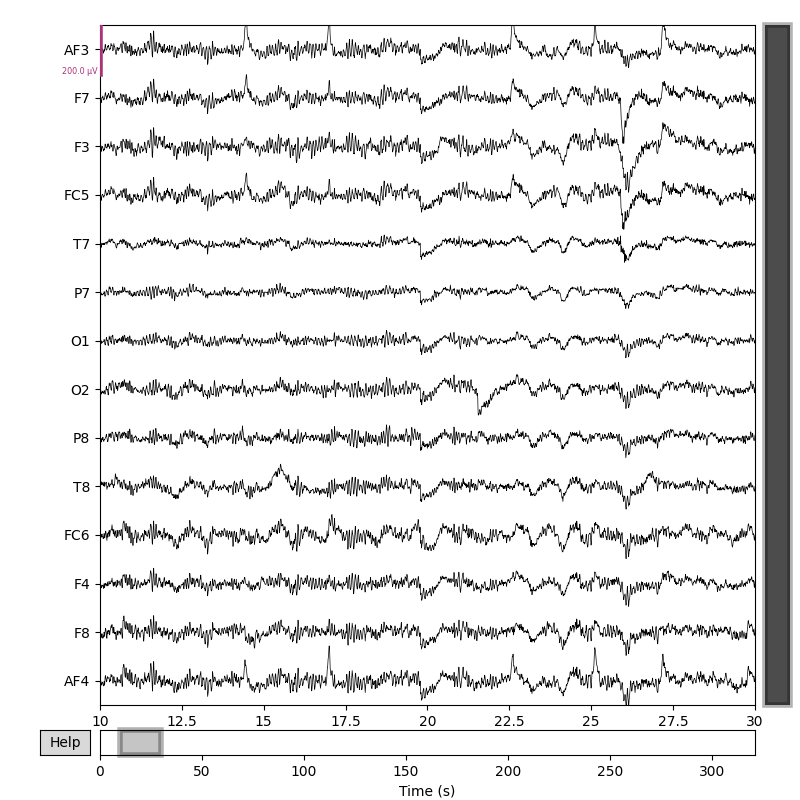
Fig 1. Raw EEG Data¶ |
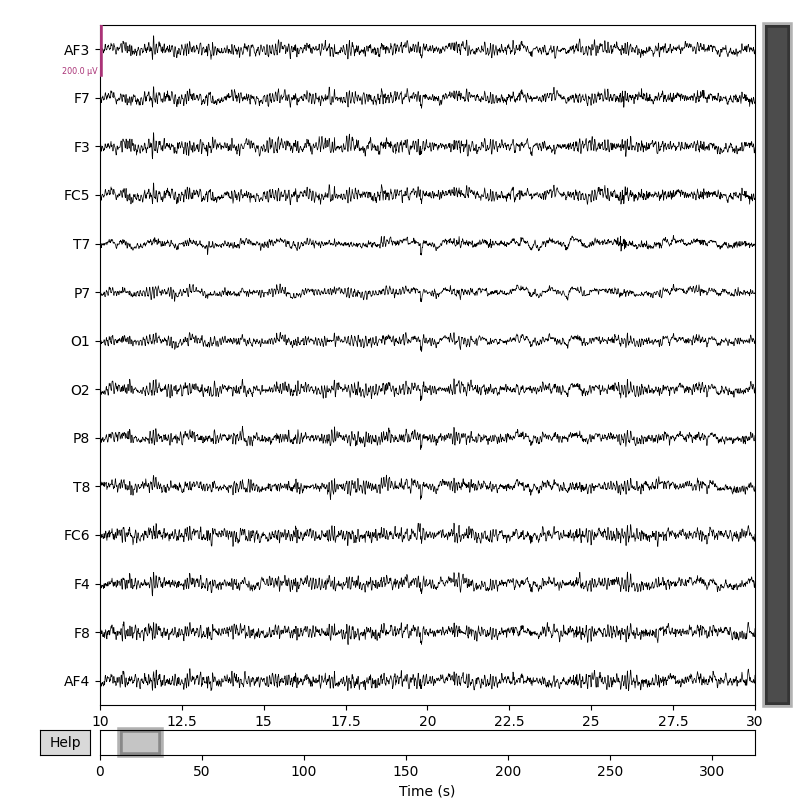
Fig 2. After Applying ATAR¶ |
5.1.3. Removing Artifacts¶
5.1.3.1. ATAR Algorithm¶
Apply ATAR on short windows of signal (single channel):
Signal is decomposed in smaller overlapping windows and reconstructed after correcting using overlap-add method.
For more details, check [1]
Operating Modes
1. Soft Thresholding
2. Elimination
3. Linear Attenuation
Computing Threshold
\(f_{\beta}(r) = k_2 \cdot exp \Big(-\beta \frac{w_{max}}{k_2} \times \frac{r}{2} \Big)\)
\(\theta_{\alpha} = f_{\beta}(r) \quad \text{if } f_{\beta}(r) \ge k_1\) otherwise \(\theta_{\alpha} = k_1\)
\(\theta_{\gamma} = g_f \times \theta_{\alpha}\) , where a default value for ‘g_f = 0.8’ For Soft-threshold
\(\theta_{\beta} = b_f \times \theta_{\alpha}\) , where a default value for ‘b_f = 2’ For Linear Attenuation
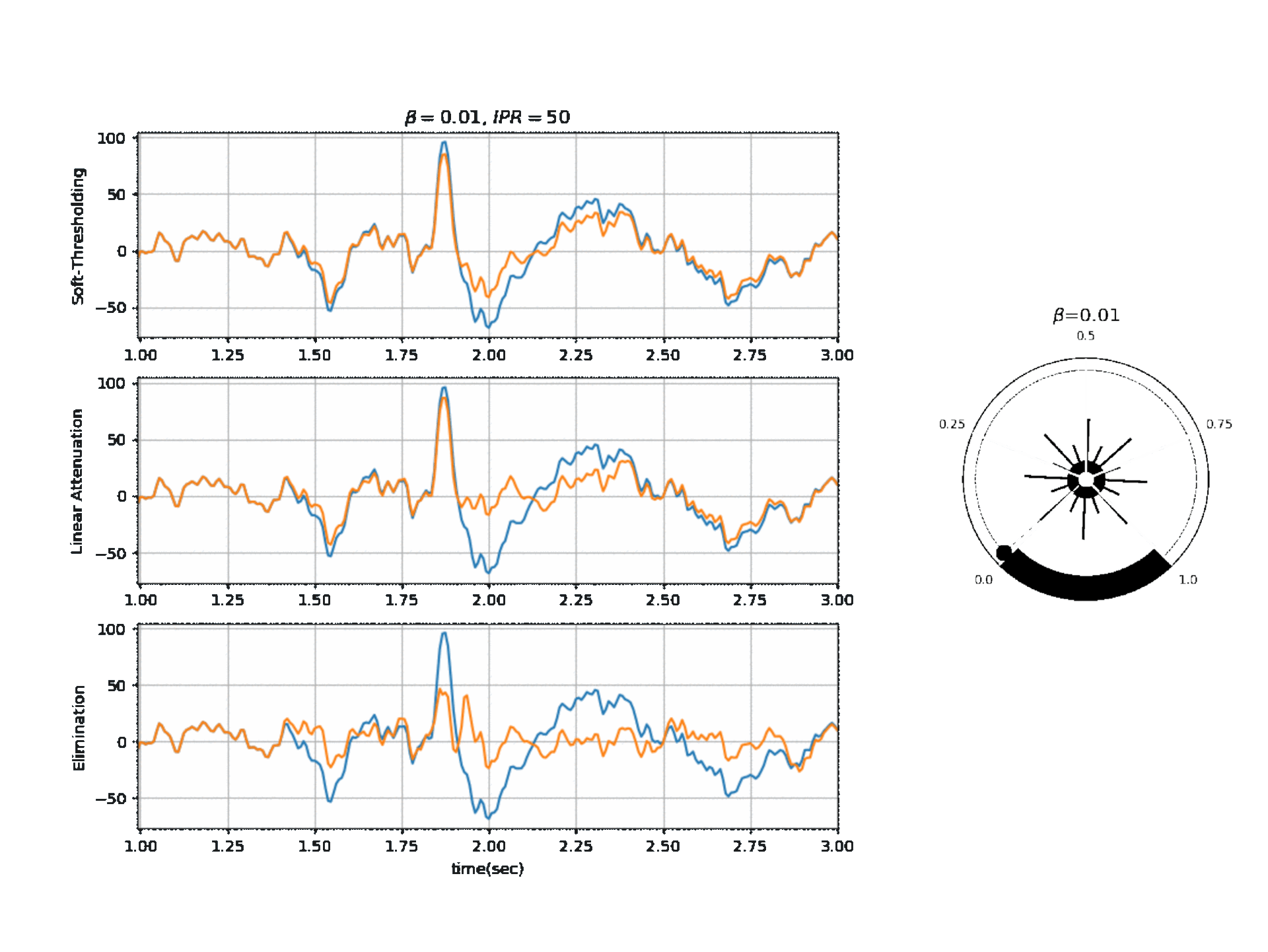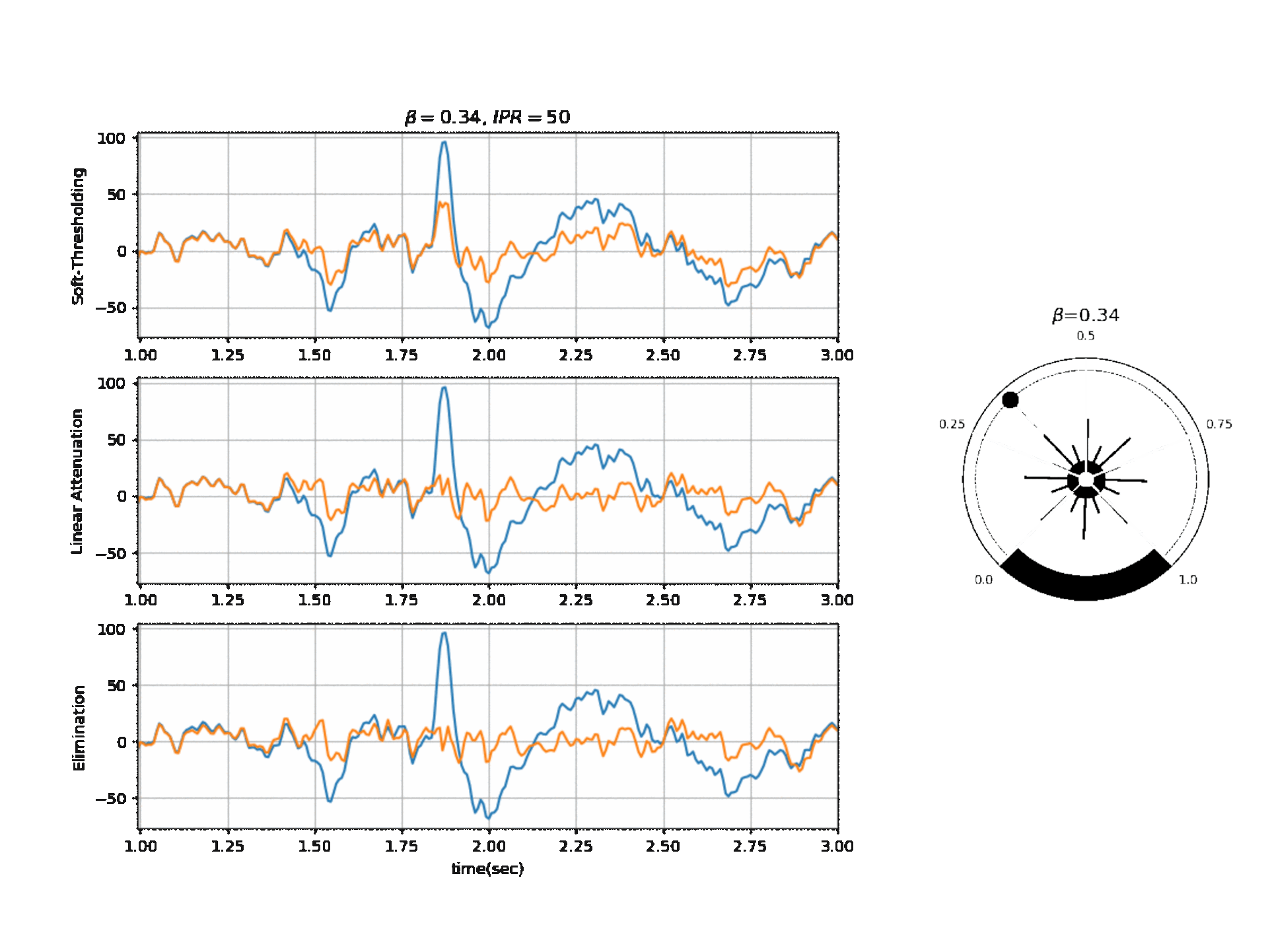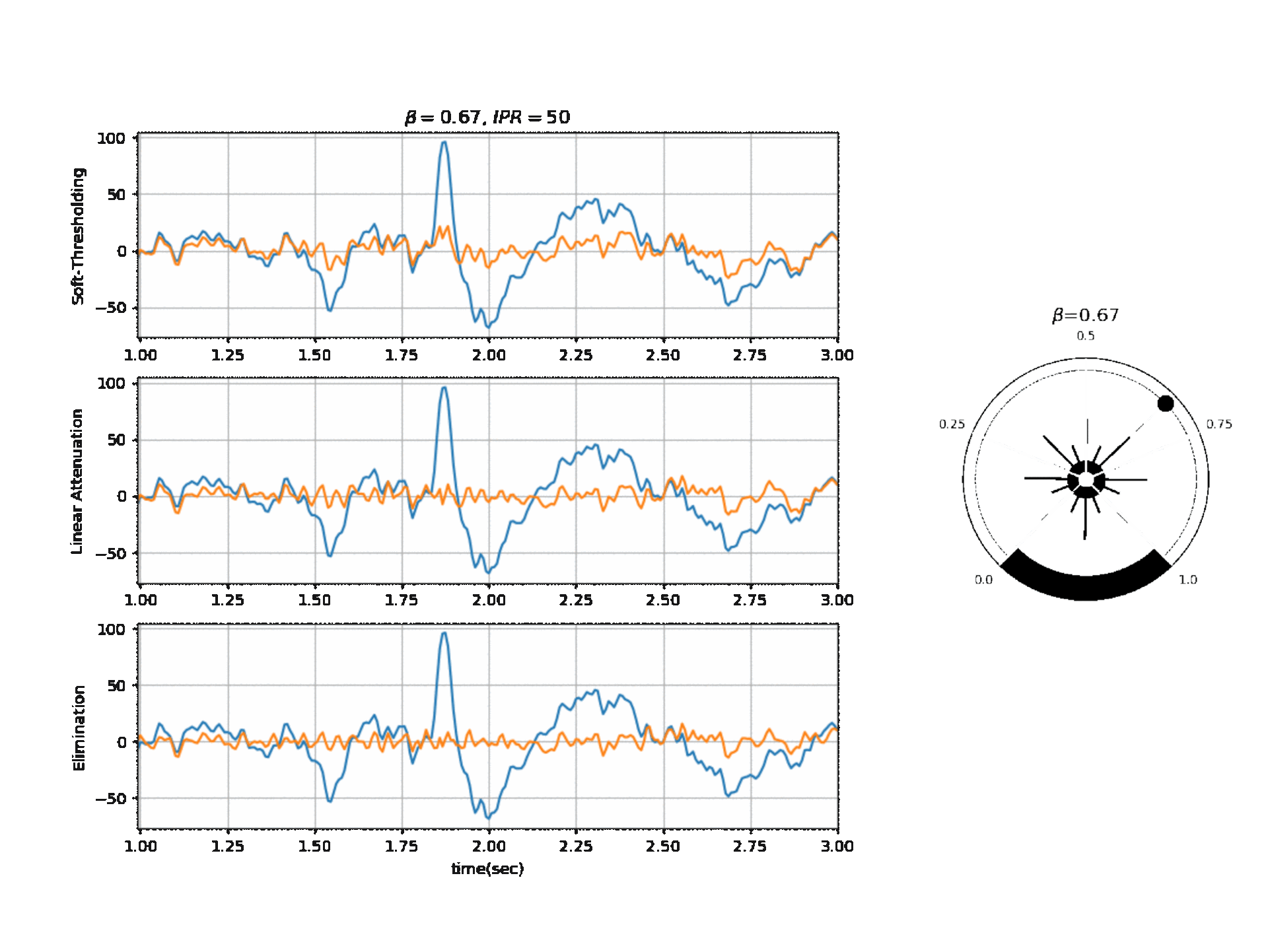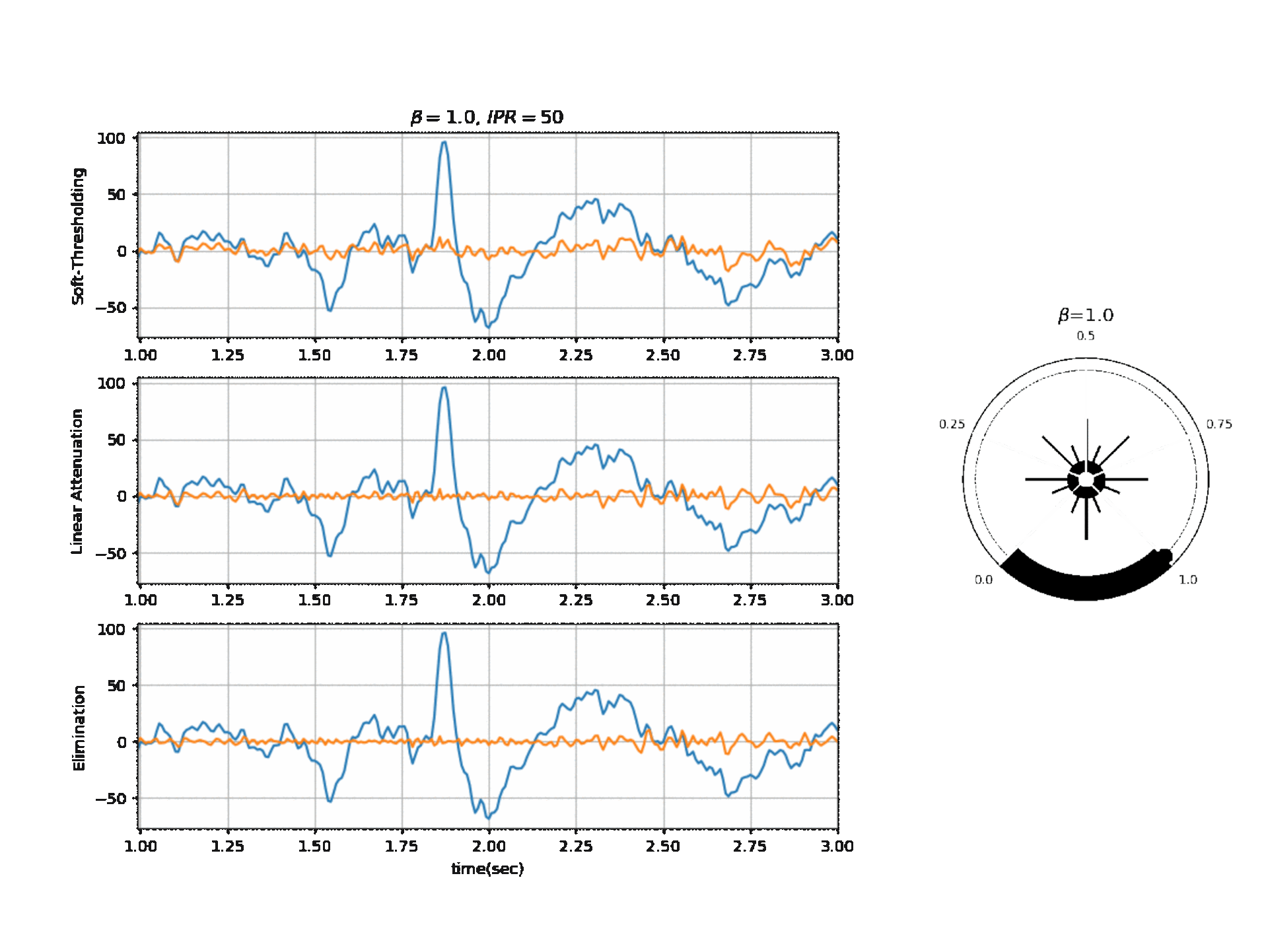
5.1.3.2. ICA Algorithm¶
ICA Filtering algorithm uses following three criteria for removing artifacts for EEG.
- Kurtosis based artifacts - mostly for motion artifacts:
- parameter
kur_thris used as threshold on kurtosis of ICs. Any IC abovekur_thr is removed. As higher kurtosis of component, more peaky it is.
- parameter
- Correlation Based Index (CBI) for eye movement artifacts:
- CBI method [1] computed the comparaision of power in prefrontal electrodes with frontal eletrodes, in IC.
A component, that stisfy the criteria, is considered as component capturing eye-blink activity and removed.
- For applying CBI method, index of prefrontal (AF - First Layer of electrodes towards frontal lobe)
and frontal lobe (F - second layer of electrodes) channels needs to be provided.
- For case of 14-channels Emotiv Epoc
ch_names = [‘AF3’,’F7’,’F3’,’FC5’,’T7’,’P7’,’O1’,’O2’,’P8’,’T8’,’FC6’,’F4’,’F8’,’AF4’]
Pre-frontal Channels =[‘AF3’,’AF4’],
Fronatal Channels = [‘F7’,’F3’,’F4’,’F8’]
- then suplied index of channels are as follow;
AF_ch_index=[0,13]F_ch_index=[1,2,11,12]
- Correlation of any IC with many EEG channels:
If any IC is correlated
corr_thr% (80%) of elecctrodes, is considered to be artifactualSimilar like CBI, except, not comparing fronatal and prefrontal but to all
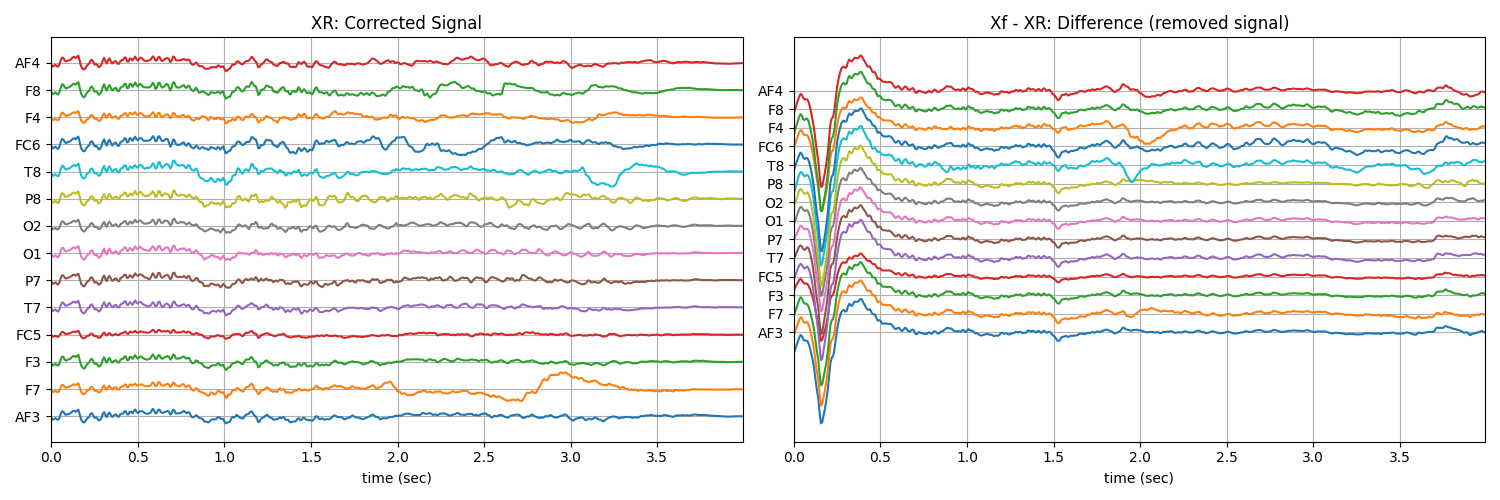
Examples:





