Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
ATAR Algorithm with MNE RAW Object¶
In this example, we show, how to apply ATAR algorithm with MNE Raw object.
We will read EDF File as MNE Raw file then extract EEG Data from it, scale and transport it for ATAR. After ATAR, we will put EEG Data back to MNE Raw object. This is perticullary useful, when all the other analysis is done in MNE Framework.
We do not show the analysis part, only, how to extract EEG from MNE Raw, apply ATAR and put it back
Same can be applied, when EEG Data file is
FIF
EDF
BDF
EGI
vhdr
Any file that can be read by mne.io.read_raw** can be used in this way to apply ATAR.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import os, re, time, glob, requests
import warnings
warnings.filterwarnings("ignore", category=DeprecationWarning)
import spkit as sp
print('spkit-version : ',sp.__version__)
import mne
spkit-version : 0.0.9.7
Download sample file¶
# Download file, if not already downloaded
path ='https://github.com/spkit/data_samples/raw/main/files/Resting_EPOCX_14Ch_Sample1.edf'
file_name = 'Resting_EPOCX_14Ch_Sample1.edf'
if not(os.path.exists(file_name)):
req = requests.get(path)
with open(file_name, 'wb') as f:
f.write(req.content)
Read EDF File with MNE Raw¶
raw = mne.io.read_raw_edf(file_name,preload=True)
print(raw.info)
fs = raw.info["sfreq"]
print('Channel Names')
sp.utils.pretty_print(raw.ch_names)
eeg_ch =['AF3', 'F7', 'F3', 'FC5', 'T7', 'P7', 'O1', 'O2', 'P8', 'T8', 'FC6', 'F4', 'F8', 'AF4']
ch_names = list(raw.ch_names)
ch_type = ['eeg' if ch in eeg_ch else 'bio' for ch in ch_names]
raw.set_channel_types(dict(zip(ch_names, ch_type)))
raw.filter(l_freq=0.5,h_freq=None, picks='eeg')
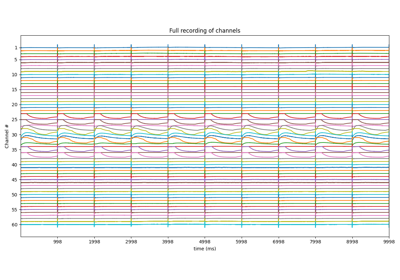
fig = raw.copy().pick('eeg').plot(scalings=dict(eeg=100e-6), duration=20, start=10)
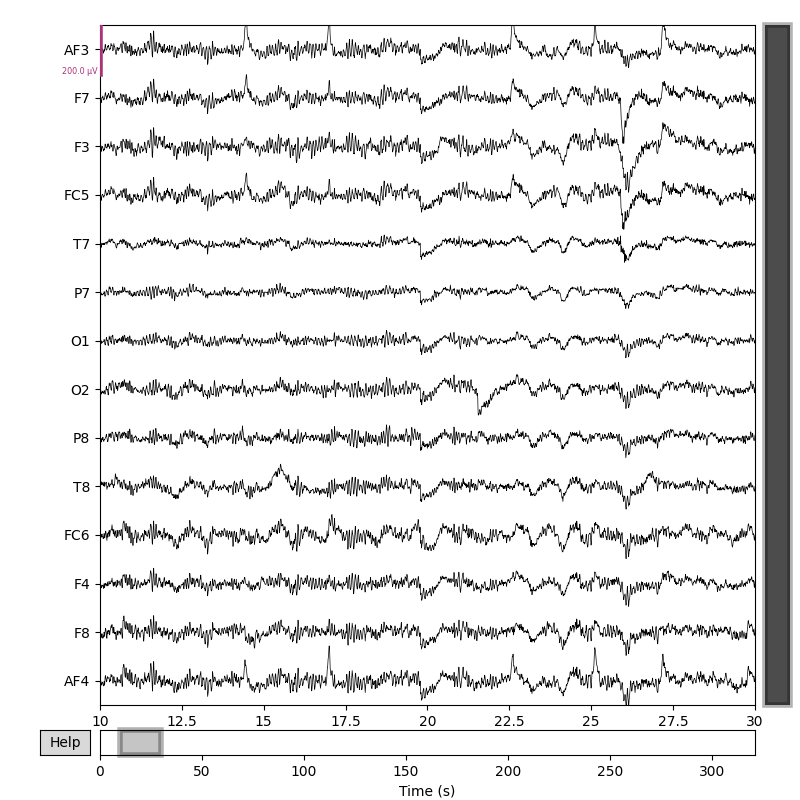
Extracting EDF parameters from /Users/nikeshbajaj/Library/CloudStorage/OneDrive-QueenMary,UniversityofLondon/Github/GIT3/Dev/SPKIT_Dev/SPKIT_DOC_7/examples/electroencephalogram/Resting_EPOCX_14Ch_Sample1.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 41087 = 0.000 ... 320.992 secs...
<Info | 8 non-empty values
bads: []
ch_names: TIME_STAMP_s, TIME_STAMP_ms, OR_TIME_STAMP_s, OR_TIME_STAMP_ms, ...
chs: 60 EEG
custom_ref_applied: False
highpass: 0.0 Hz
lowpass: 64.0 Hz
meas_date: 2022-11-03 16:34:32 UTC
nchan: 60
projs: []
sfreq: 128.0 Hz
subject_info: 1 item (dict)
>
Channel Names
0 TIME_STAMP_s | 1 TIME_STAMP_ms | 2 OR_TIME_STAMP_s
3 OR_TIME_STAMP_ms | 4 COUNTER | 5 INTERPOLATED
6 AF3 | 7 F7 | 8 F3
9 FC5 | 10 T7 | 11 P7
12 O1 | 13 O2 | 14 P8
15 T8 | 16 FC6 | 17 F4
18 F8 | 19 AF4 | 20 RAW_CQ
21 BATTERY | 22 BATTERY_PERCENT | 23 MarkerIndex
24 MarkerType | 25 MarkerValueInt | 26 MARKER_HARDWARE
27 CQ_AF3 | 28 CQ_F7 | 29 CQ_F3
30 CQ_FC5 | 31 CQ_T7 | 32 CQ_P7
33 CQ_O1 | 34 CQ_O2 | 35 CQ_P8
36 CQ_T8 | 37 CQ_FC6 | 38 CQ_F4
39 CQ_F8 | 40 CQ_AF4 | 41 CQ_OVERALL
42 EQ_SampleRateQua| 43 EQ_OVERALL | 44 EQ_AF3
45 EQ_F7 | 46 EQ_F3 | 47 EQ_FC5
48 EQ_T7 | 49 EQ_P7 | 50 EQ_O1
51 EQ_O2 | 52 EQ_P8 | 53 EQ_T8
54 EQ_FC6 | 55 EQ_F4 | 56 EQ_F8
57 EQ_AF4 | 58 CQ_CMS | 59 CQ_DRL
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 0.5 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.50
- Lower transition bandwidth: 0.50 Hz (-6 dB cutoff frequency: 0.25 Hz)
- Filter length: 845 samples (6.602 s)
Using matplotlib as 2D backend.
Extract EEG and Apply ATAR¶
X = raw.get_data()
#EEG Channels
XE = X[6:20].copy()
print(XE.shape, XE.min(), XE.max())
# Scale in uV (order of 100s) and transpose (channle at axis =1)
XE = XE.T*1e6
# ATAR
XR = sp.eeg.ATAR(XE,wv='db3',winsize=128, thr_method='ipr', OptMode='elim',beta=0.1,k2=100,verbose=1,use_joblib=False)
# Scale and transpose back
XR = XR.T*1e-6
(14, 41088) -0.0003016368415400783 0.00046421187258672094
Putting back EEG to Original Raw MNE¶
raw1 = raw.copy()
Xi = raw1.get_data()
Xi[6:20] = XR
raw1._data = Xi
Plot for comparison¶
fig = raw1.copy().pick('eeg').plot(scalings=dict(eeg=100e-6), duration=20, start=10)
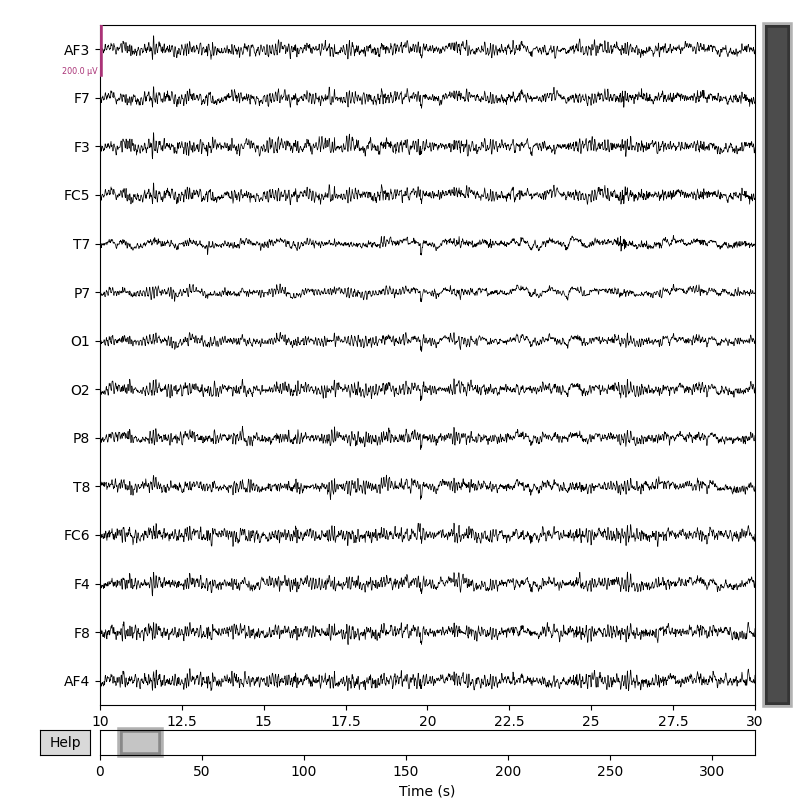
Total running time of the script: (0 minutes 5.141 seconds)
Related examples
ATAR: Automatic and Tunable Artifact Removal Algorithm