spkit.eeg.ATAR¶
- spkit.eeg.ATAR(X, wv='db3', winsize=128, thr_method='ipr', OptMode='soft', IPR=[25, 75], beta=0.1, k1=10, k2=100, est_wmax=100, theta_a=inf, bf=2, gf=0.8, wpd_mode='symmetric', wpd_maxlevel=None, recons_method='atar-style', window=['hamming', True], hopesize=None, packetwise=False, WPD=True, lvl=[], use_joblib=False, verbose=False, bound_warn=True, **kwargs)¶
ATAR - Automatic and Tunable Artifact Removal Algorithm
ATAR: - Automatic and Tunable Artifact Removal Algorithm
Apply ATAR on short windows of signal (single channel):
Signal is decomposed in smaller overlapping windows and reconstructed after correcting using overlap-add method.
For more details, check [1]
Operating Modes
1. Soft Thresholding
\[ \begin{align}\begin{aligned}\lambda_s (w) &= w & \quad \text{if } |w|<\theta_{\gamma}\\ &= \theta_{\alpha} \frac{1 - e^{\alpha w}}{1 + e^{\alpha w}} & \quad \text{otherwise}\end{aligned}\end{align} \]2. Elimination
\[ \begin{align}\begin{aligned}\lambda_e (w) &= w & \quad \text{if } |w| \le \theta_{\alpha}\\ &= 0 & \quad \text{otherwise}\end{aligned}\end{align} \]3. Linear Attenuation
\[ \begin{align}\begin{aligned}\lambda_a (w) &= w & \quad \text{if } |w| \le \theta_{\alpha}\\ &= sgn(w) \theta_{\alpha} \Big( 1 - \frac{|w| - \theta_{\alpha}}{\theta_{\beta}-\theta_{\alpha}} \Big) & \quad \text{if } \theta_{\alpha} < |w| \le \theta_{\beta}\\ &= 0 & \quad \text{otherwise}\end{aligned}\end{align} \]Computing Threshold
\(f_{\beta}(r) = k_2 \cdot exp \Big(-\beta \frac{w_{max}}{k_2} \times \frac{r}{2} \Big)\)
\(\theta_{\alpha} = f_{\beta}(r) \quad \text{if } f_{\beta}(r) \ge k_1\) otherwise \(\theta_{\alpha} = k_1\)
\(\theta_{\gamma} = g_f \times \theta_{\alpha}\) , where a default value for ‘g_f = 0.8’ For Soft-threshold
\(\theta_{\beta} = b_f \times \theta_{\alpha}\) , where a default value for ‘b_f = 2’ For Linear Attenuation
- Parameters:
- x1d-array
input single-channel signal of shape (n,)
- wv = str, ‘db3’(default) wavelet
- one of wavelet family {‘db3’…..’db38’, ‘sym2…..sym20’,
‘coif1…..coif17’, ‘bior1.1….bior6.8’,
‘rbio1.1…rbio6.8’, ‘dmey’}
- winsize: int, deafult=128
window size to apply ATAR
- hopesize: int, None, default=None,
overlap shift for next window
if None, hopesize=winsize//2
only used when recons_method={‘atar-style’,’atar’,’custom’}
- Operating modes
- OptMode: str, {‘soft’,’elim’,’linAtten’}, default ‘soft’
Operating Modes:
soft: soft-thresholding (need theta_a, and theta_g)
elim: Elimination mode (need theta_a)
- linAtten: Linear Attenuation mode (need theta_a and theta_b)
given
bfandgf(default bf = 2, gf=0.8)- where:-
theta_b = bf*theta_a – used for Linear Attenuation
theta_g = gf*theta_a – used for Soft thresholding
- *Threshold Computation method*
- thr_method: str, {‘ipr’, None}
Computing method for threshold
if ‘ipr’ ‘theta_a’ is computed as per ATAR’s approach
if None, passed value of ‘theta_a’ is used along with (bf, gf)
- IPR: list of two default=[25,75]
To compute Interpercentile range r
e.g. [10,90], [30,70]
Higher the range is, threshold is more affected by outliers
Interpercentile range r is mapped to threshold, using
beta,k1andk2
- beta: float (0,1] default=0.1
beta as tuning parameter for threshold
higher the value, more aggressive is the algorithm to remove artifacts
- (k1,k2) :scalars, defualt (10,100)
lower and upper bounds on the threshold value
should be of same order as signal magnitude.
if signal is in volts, values will be of order 1e-6, then k1 and k2 should be around those order
defult values k1=10, k2=100 are chosen for signal with unit of microVolt (uV), of order of 100s
Warning
k1,k2 bounds if k2 is very high (e.g. 100) and signal amplitude is in 1e-3, 1e-6, ATAR will have no effect on the signal, as theshold to identify and remove artifacts will be so high.
- est_wmax: int, default=100
est_wmax is the value in the expression (15 in paper) or above Computing Threshold equations
w_max
- *Wavelet Decomposition*
- wpd_mode: str, default ‘symmetric’
- one of the {‘zero’, ‘constant’, ‘symmetric’, ‘periodic’,
‘smooth’, ‘periodization’}
- wpd_maxlevel: int, defualt=None
maximum number of levels for decomposition,
if None, max possible number of level are used.
- use_joblib: bool, default=False
If True, joblib is used for parallel processing of the channels
- verbose: int
vebosity level
- Experimental Settings
Note
NOT RECOMMONEDED TO CHANGE Following parameters are experimental, they are not recommonded to change, and leave as default
- WPD: bool, default=True
if False Wavelet Transform is used for decompisiton of signal, else Wavelet Packet Decomposition is used
- packetwise: bool, deafult=False
if True, threshold is computed and applied to each packet independently.
- lvl: list defualt=[]
if provided, ATAR is applied to provided level numbers only.
- recons_method: str deafult=’atar-style’
reconstruction method after applying atar to each window.
one of {‘atar-style’, ‘atar’, ‘custom’, ‘HamWin’,’Hamming’}
Note
NOT RECOMMONEDED TO CHANGE KEEP IT TO DEFAULT
- window: list of two default=[‘hamming’,True]
window function, and if windowing is applied before or after atar
- Returns:
- XR: corrected signal of same shape as input X
See also
ATAR_1ChATAR for single channel
ATAR_mChATAR for multiple channel
ICA_filteringICA based Artifact Removal Algorithm
References
[1]Bajaj, Nikesh, et al. “Automatic and tunable algorithm for EEG artifact removal using wavelet decomposition with applications in predictive modeling during auditory tasks.” Biomedical Signal Processing and Control 55 (2020): 101624. https://doi.org/10.1016/j.bspc.2019.101624
Examples
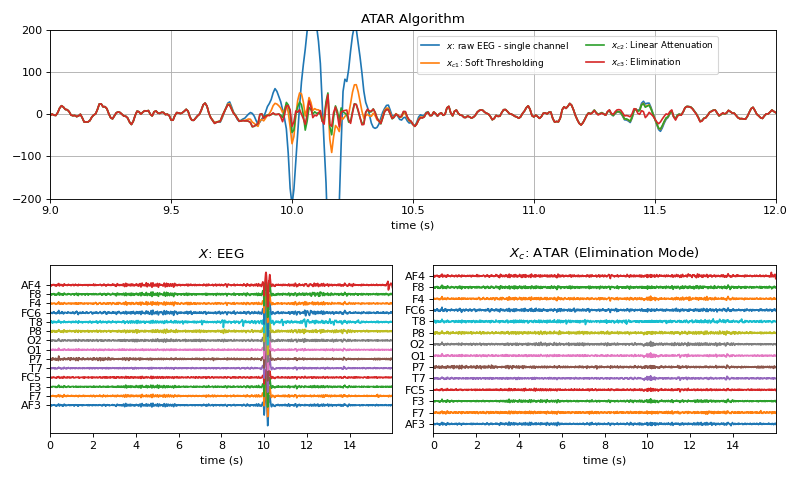
#sp.eeg.ATAR import numpy as np import matplotlib.pyplot as plt import spkit as sp X,fs, ch_names = sp.data.eeg_sample_14ch() X = sp.filterDC_sGolay(X, window_length=fs//3+1) t = np.arange(X.shape[0])/fs # Single Channel x = X[:,0] xc1 = sp.eeg.ATAR(x,wv='db3', winsize=128, thr_method='ipr',beta=0.1, k1=10, k2=100,OptMode='soft',verbose=1) xc2 = sp.eeg.ATAR(x,wv='db3', winsize=128, thr_method='ipr',beta=0.1, k1=10, k2=100,OptMode='linAtten',verbose=1) xc3 = sp.eeg.ATAR(x,wv='db3', winsize=128, thr_method='ipr',beta=0.1, k1=10, k2=100,OptMode='elim',verbose=1) # Multi-Channel Xc = sp.eeg.ATAR(X,wv='db3', winsize=128, thr_method='ipr',beta=0.2, k1=10, k2=100,OptMode='elim') sep=200 plt.figure(figsize=(10,6)) plt.subplot(211) plt.plot(t,x, label='$x$: raw EEG - single channel') plt.plot(t,xc1,label=r'$x_{c1}$: Soft Thresholding') plt.plot(t,xc2,label=r'$x_{c2}$: Linear Attenuation') plt.plot(t,xc3,label=r'$x_{c3}$: Elimination') plt.xlim([9,12]) plt.ylim([-200,200]) plt.legend(bbox_to_anchor=(0.5,0.99),ncol=2,fontsize=8) plt.grid() plt.title(r'ATAR Algorithm') plt.xlabel('time (s)') plt.subplot(223) plt.plot(t,X+np.arange(X.shape[1])*sep) plt.xlim([t[0],t[-1]]) plt.yticks(np.arange(X.shape[1])*sep,ch_names) plt.title(r'$X$: EEG') plt.xlabel('time (s)') plt.subplot(224) plt.plot(t,Xc+np.arange(14)*sep) plt.xlim([t[0],t[-1]]) plt.title(r'$X_c$: ATAR (Elimination Mode)') plt.yticks(np.arange(X.shape[1])*sep,ch_names) plt.xlabel('time (s)') plt.tight_layout() plt.show()