spkit.eeg.ICA_filtering¶
- spkit.eeg.ICA_filtering(X, winsize=128, ICA_method='extended-infomax', kur_thr=2, corr_thr=0.8, AF_ch_index=None, F_ch_index=None, verbose=True, window=['hamming', True], hopesize=None, winMeth='custom')¶
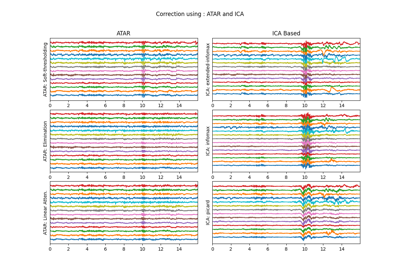
Aftifact Removal using ICA
Aftifact Removal using ICA
ICA Filtering algorithm uses following three criteria for removing artifacts for EEG.
- Kurtosis based artifacts - mostly for motion artifacts:
parameter
kur_thris used as threshold on kurtosis of ICs. Any IC abovekur_thris removed. As higher kurtosis of component, more peaky it is.
- Correlation Based Index (CBI) for eye movement artifacts:
CBI method [R3cb126c15dac-1] computed the comparaision of power in prefrontal electrodes with frontal eletrodes, in IC. A component, that stisfy the criteria, is considered as component capturing eye-blink activity and removed.
For applying CBI method, index of prefrontal (AF - First Layer of electrodes towards frontal lobe) and frontal lobe (F - second layer of electrodes) channels needs to be provided.
- For case of 14-channels Emotiv Epoc
ch_names = [‘AF3’,’F7’,’F3’,’FC5’,’T7’,’P7’,’O1’,’O2’,’P8’,’T8’,’FC6’,’F4’,’F8’,’AF4’]
Pre-frontal Channels =[‘AF3’,’AF4’],
Fronatal Channels = [‘F7’,’F3’,’F4’,’F8’]
- then suplied index of channels are as follow;
AF_ch_index=[0,13]F_ch_index=[1,2,11,12]
- Correlation of any IC with many EEG channels:
If any IC is correlated
corr_thr% (80%) of elecctrodes, is considered to be artifactualSimilar like CBI, except, not comparing fronatal and prefrontal but to all
- Parameters:
- X: array,(n,ch)
input signal (n,ch) with n samples and ch channels
- winsize: int, default=128
window size to process, if None, entire signal is used at once
- ICAMed = {‘fastICA’,’infomax’,’extended-infomax’,’picard’}
method of ICA Decomposition
- kur_thr: + scalar, default=2
threshold on kurtosis of IC component, as per (1) criteria (see above)
Note
CBI Method if
AF_ch_indexorF_ch_indexis None, CBI is not applied.- AF_ch_index: list, None
(AF - First Layer of electrodes towards frontal lobe)
example [0,13], as for [‘AF3’,’AF4’] for Emotiv 14 channel data
check above details for suplying accurate indices, as per your data
Changed in version 0.0.9.7: Default is set to None, instead of [0,13]
- F_ch_index: list, None
(F - second layer of electrodes)
example [1,2,11,12], as for [‘F7’,’F3’,’F4’,’F8’] for Emotiv 14 channel data
check above details for suplying accurate indices, as per your data
Changed in version 0.0.9.7: Default is set to None, instead of [1,2,11,12]
- corr_thr: float [0, 1] (deafult 0.8)
threshold to consider correlation, higher the value less IC are removed and vise-versa
if None, this is not applied
Note
Criteria (3) if
corr_thris None then Criteria (3) is not applied.
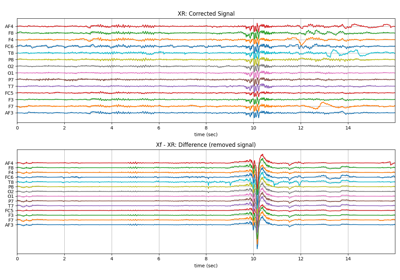
- Returns:
- XR: array,(n,ch)
Filtered signal, same size as X, (n,ch)
See also
References
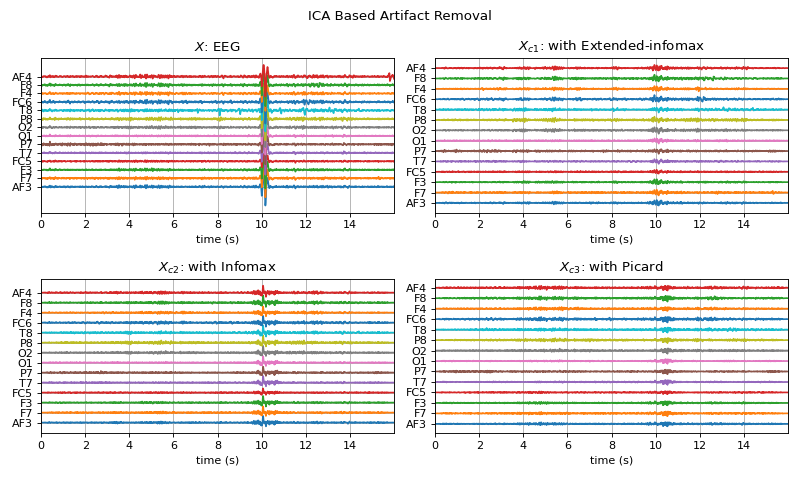
Examples
#sp.eeg.ICA_filtering import numpy as np import matplotlib.pyplot as plt import spkit as sp X,fs, ch_names = sp.data.eeg_sample_14ch() X = sp.filterDC_sGolay(X, window_length=fs//3+1) t = np.arange(X.shape[0])/fs Xc1 = sp.eeg.ICA_filtering(X.copy(),winsize=128, ICA_method='extended-infomax',kur_thr=2,corr_thr=0.8, AF_ch_index = [0,13] ,F_ch_index=[1,2,11,12]) Xc2 = sp.eeg.ICA_filtering(X.copy(),winsize=128, ICA_method='infomax',kur_thr=2,corr_thr=0.8, AF_ch_index = [0,13] ,F_ch_index=[1,2,11,12]) Xc3 = sp.eeg.ICA_filtering(X.copy(),winsize=128, ICA_method='picard',kur_thr=2,corr_thr=0.8, AF_ch_index = [0,13] ,F_ch_index=[1,2,11,12]) sep=200 plt.figure(figsize=(10,6)) plt.subplot(221) plt.plot(t,X+np.arange(X.shape[1])*sep) plt.xlim([t[0],t[-1]]) plt.yticks(np.arange(X.shape[1])*sep,ch_names) plt.title(r'$X$: EEG') plt.xlabel('time (s)') plt.grid() plt.subplot(222) plt.plot(t,Xc1+np.arange(14)*sep) plt.xlim([t[0],t[-1]]) plt.title(r'$X_{c1}$: with Extended-infomax') plt.yticks(np.arange(Xc1.shape[1])*sep,ch_names) plt.xlabel('time (s)') plt.grid() plt.subplot(223) plt.plot(t,Xc2+np.arange(14)*sep) plt.xlim([t[0],t[-1]]) plt.title(r'$X_{c2}$: with Infomax') plt.yticks(np.arange(Xc2.shape[1])*sep,ch_names) plt.xlabel('time (s)') plt.grid() plt.subplot(224) plt.plot(t,Xc3+np.arange(14)*sep) plt.xlim([t[0],t[-1]]) plt.title(r'$X_{c3}$: with Picard') plt.yticks(np.arange(Xc3.shape[1])*sep,ch_names) plt.xlabel('time (s)') plt.grid() plt.suptitle(r'ICA Based Artifact Removal') plt.tight_layout() plt.show()