spkit.data.optical_sample¶
- spkit.data.optical_sample(sample=1, species=None)¶
Load sample(s) of optical mapping signals
- Parameters:
- sampleint, {1,2,3,-1}
sample number
if -1, return all three samples
- species: str, default=None, {None, ‘rabbit’}
data of species
for ‘rabbit’, data of two cameras
- Returns:
- Xarray,
shape (n,) or (n,3) if species= None
shape (n,2) or shape (n,6) for species=’rabbit’,
- fsint
See also
eda_sampleElectrodermal activity (EDA)
gsr_sampleGalvanic Skin Response (GSR)
eeg_sample_14chElectroencephalography (EEG) - 14-channel
eeg_sample_1chElectroencephalography (EEG) - 1-channel
eeg_sample_artifactElectroencephalography (EEG) processed
ecg_sample_12leadsElectrocardiogram (ECG) - 12-leads
ecg_sampleElectrocardiogram (ECG) - 1-lead
ppg_samplePhotoplethysmogram (PPG)
egm_sampleElectrogram (EGM)
Examples
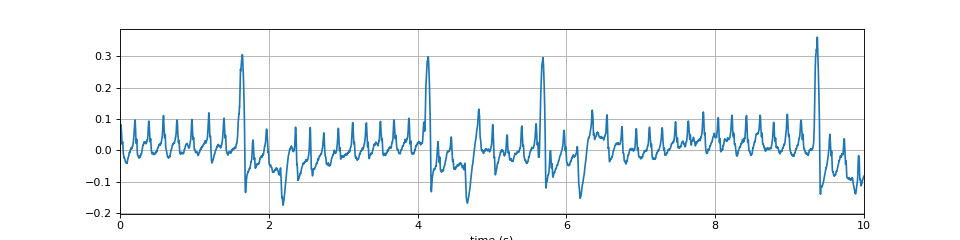
import numpy as np import matplotlib.pyplot as plt import spkit as sp x,fs = sp.data.optical_sample(sample=1) t = np.arange(len(x))/fs plt.figure(figsize=(12,3)) plt.plot(t,x) plt.xlim([t[0],t[-1]]) plt.xlabel('time (s)') plt.grid() plt.show()
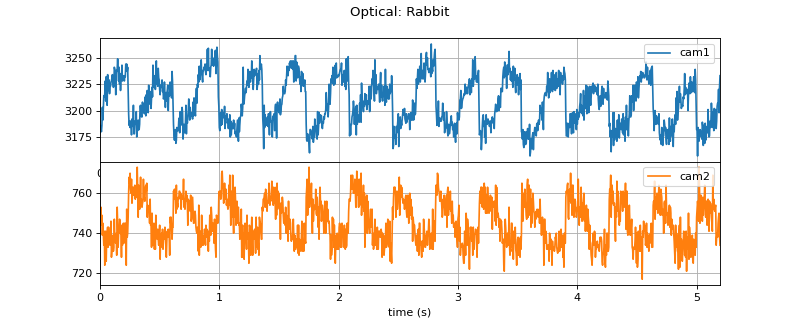
#sp.data.optical_sample import numpy as np import matplotlib.pyplot as plt import spkit as sp X,fs = sp.data.optical_sample(sample=1,species='rabbit') t = np.arange(X.shape[0])/fs plt.figure(figsize=(10,4)) plt.subplot(211) plt.plot(t,X[:,0],color='C0',label='cam1') plt.xlim([t[0],t[-1]]) plt.grid() plt.legend(loc=1) plt.subplot(212) plt.plot(t,X[:,1],color='C1',label='cam2') plt.xlim([t[0],t[-1]]) plt.grid() plt.legend(loc=1) plt.xlabel('time (s)') plt.subplots_adjust(hspace=0) plt.suptitle('Optical: Rabbit') plt.show()