spkit.data.eda_sample¶
- spkit.data.eda_sample(sample=1)¶
Load sample(s) of Electrodermal activity (EDA)
Widely known as Electrodermal activity (EDA)
Sample is recorded using copper plats at lower sampling rate (128)
Sample is part of PhyAAt Dataset [2]
- Parameters:
- sampleint, {1,2,-1}
sample number
if -1, return all two samples
- Returns:
- Xarray, (n,) or (n,2)
- fsint
See also
gsr_sampleGalvanic Skin Response (GSR)
eeg_sample_14chElectroencephalography (EEG) - 14-channel
eeg_sample_1chElectroencephalography (EEG) - 1-channel
eeg_sample_artifactElectroencephalography (EEG) processed
ecg_sample_12leadsElectrocardiogram (ECG) - 12-leads
ecg_sampleElectrocardiogram (ECG) - 1-lead
optical_sampleOptical Mapped Signal
ppg_samplePhotoplethysmogram (PPG)
egm_sampleElectrogram (EGM)
References
[1] wikipedia - https://en.wikipedia.org/wiki/Electrodermal_activity
[2] PhyAAt Dataset - https://phyaat.github.io/
Examples
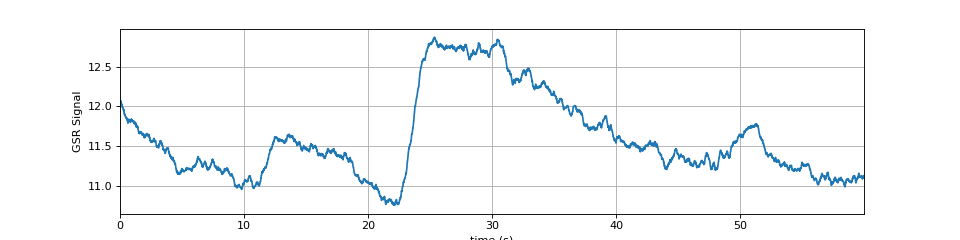
#sp.data.eda_sample #Example 1 import numpy as np import matplotlib.pyplot as plt import spkit as sp x,fs = sp.data.eda_sample(sample=1) x = x[:int(fs*60)] t = np.arange(len(x))/fs plt.figure(figsize=(12,3)) plt.plot(t,x) plt.xlim([t[0],t[-1]]) plt.xlabel('time (s)') plt.ylabel('GSR Signal') plt.grid() plt.show()
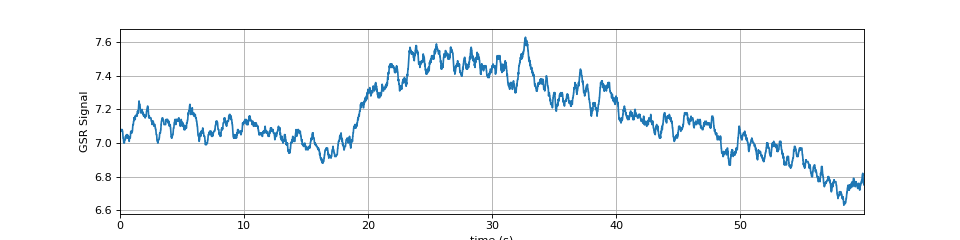
############################ #sp.data.gsr_sample #Example 2 import numpy as np import matplotlib.pyplot as plt import spkit as sp x,fs = sp.data.eda_sample(sample=2) x = x[:int(fs*60)] t = np.arange(len(x))/fs plt.figure(figsize=(12,3)) plt.plot(t,x) plt.xlim([t[0],t[-1]]) plt.xlabel('time (s)') plt.ylabel('GSR Signal') plt.grid() plt.show()