spkit.conv2d_nan¶
- spkit.conv2d_nan(x, kernel, boundary='fill', fillvalue=0, denormalise_ker=True)¶
2D Convolution with NaN values
2D Convolution with NaN values
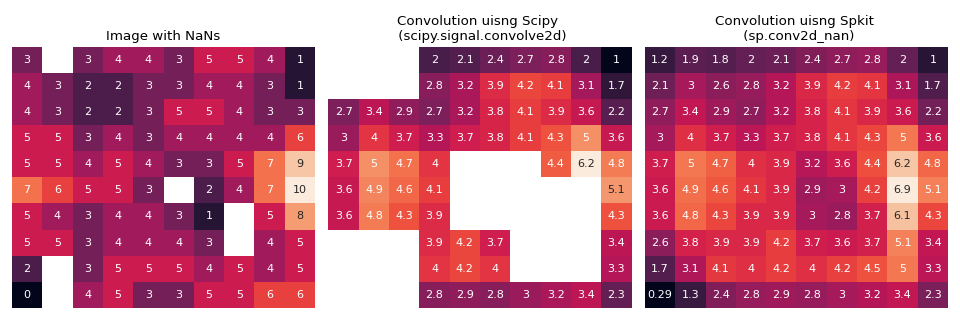
In conventional Convolution funtions, if any of the value in input x or in kernel is NaN (np.nan), then NaN values are propogated and corrupt other values too.
To avoid this, this funtion does the convolution in same fashion as conventional except, it allows NaN values to exists in input, without propogating their effect. It even replaces them.
While computating, it simply ignores the NaN value, as if does not exist, and adjust the computation accordingly.
If No NaN value exist, result is same as conventional convolution
- Parameters:
- x: 2D-array with NaN values.
- kernel: a 2D kernel
to use for convolution
IMPORTANT NOTE: kernel passed should NOT be normalised. If normalised kernel is used, the results will be very different than conventional convolution.
- For example:
kernel_unnorm = np.ones([3,3]) kernel_norm = np.ones([3,3])/9
kernel_unnorm should be passed, not kernel_norm.
To de-normalise a kernel, used
denorm_kernelor set denormalise_ker=True
- denormalise_ker: bool, default=True
If True, first de-normalise kernel
- boundarystr {‘fill’,’constant’ ,’wrap’, ‘symm’,’symmetric’,’reflect’,}, optional
A flag indicating how to handle boundaries:
fillorconstantpad input arrays with fillvalue. (default)
wrapcircular boundary conditions.
symmor ‘symmetric’symmetrical boundary conditions.
reflectreflecting boundary conditions.
- fillvalue: scalar, optional
Value to fill pad input arrays with. Default is np.nan,
- Returns:
- y: 2D-arrray of same size as input x with no NaN propogation
See also
Examples
>>> #sp.conv2d_nan >>> import numpy as np >>> import matplotlib.pyplot as plt >>> import seaborn as sns >>> import scipy >>> import spkit as sp >>> seed = 2 >>> I = (sp.create_signal_2d(n=10,seed=seed,sg_winlen=3)*10).round(0) >>> np.random.seed(seed) >>> r = np.random.rand(*I.shape) >>> np.random.seed(None) >>> I[r<0.05] = np.nan >>> kernel = np.ones([3,3])/9 >>> I_scipy = scipy.signal.convolve2d(I.copy(), kernel,mode='same', boundary='fill',fillvalue=0) >>> I_spkit = sp.conv2d_nan(I.copy(), kernel, boundary='fill',fillvalue=0) >>> plt.figure(figsize=(12,4)) >>> plt.subplot(131) >>> sns.heatmap(I, annot=True,cbar=False,xticklabels='', yticklabels='') >>> plt.title('Image with NaNs') >>> plt.subplot(132) >>> sns.heatmap(I_scipy, annot=True,cbar=False,xticklabels='', yticklabels='') >>> plt.title('Convolution uisng Scipy \n (scipy.signal.convolve2d)') >>> plt.subplot(133) >>> sns.heatmap(I_spkit, annot=True,cbar=False,xticklabels='', yticklabels='') >>> plt.title('Convolution uisng Spkit \n (sp.conv2d_nan)') >>> plt.tight_layout() >>> plt.show()