spkit.ml.LogisticRegression¶
- class spkit.ml.LogisticRegression(alpha=0.01, lambd=0, polyfit=False, degree=3, FeatureNormalize=False, penalty='l2', tol=0.01, rho=0.9, C=1.0, fit_intercept=True)¶
Logistic Regression
fitting a linear model
For binary
\[y_p = sigmoid(X \times W + b)\]For multiclass
\[y_p = softmax(X \times W + b)\]Optimizing \(W\) and \(b\), using gradient decent and regularize with ‘l1’,’l2’ or ‘elastic-net’ penalty
- Parameters:
- fit_intercept: bool, default=True
if use intercept (b) to fit, if false, b = 0
- alpha: scalar,
Learning rate, the rate to update the weights at each iteration
- penalty: str, {‘l1’, ‘l2, ‘elastic-net’, ‘none’}, default=’l2’
- regularization:
‘l1’ –> lambd*|W|,
‘l2’ –> lambd*|W|**2
‘elastic-net’ - lambd*(rho*|W|^2 + (1-rho)*|W|))
- lambdscalar, default =0
penalty value ()
- rhoscalar
used for elastic-net penalty
- tolfloat, default=1e-4
Tolerance for stopping criteria.
- polyfit: bool, default=False
if polynomial features to be used,
- degree: int,default=2
degree of polynomial features, used if polyfit is true,
- FeatureNormalize: bool, default=False
- if true, before fitting and after polyfit, features are normalized
and computed mean and std is saved to be used while prediction
- Attributes:
- nclass: ndarray of shape (n_classes, )
A list of class labels known to the classifier.
- W: ndarray of shape (1, n_features) or (n_classes, n_features)
Coefficient of the features in the decision function.
- b: intercept weight
intercept weight
See also
Examples
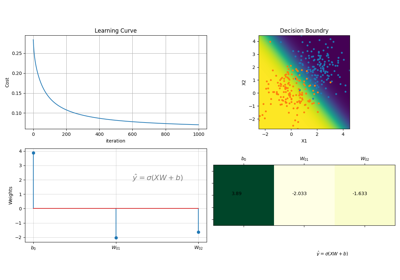
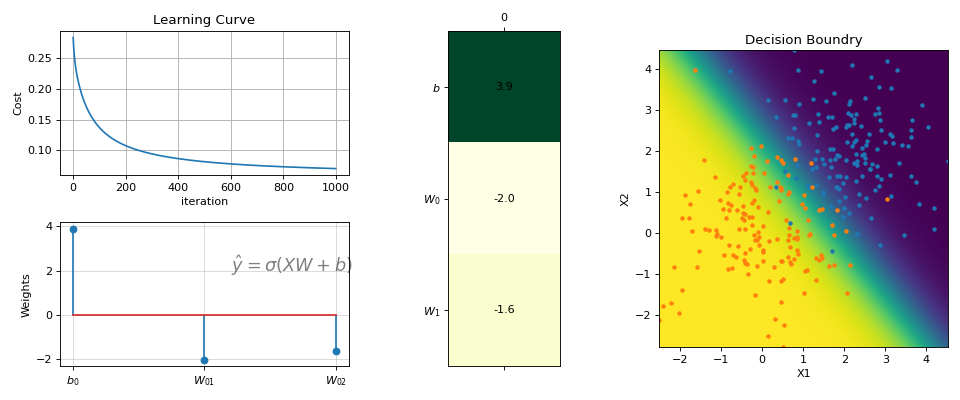
#sp.ml.LogisticRegression import numpy as np import matplotlib.pyplot as plt import spkit as sp from spkit.ml import LogisticRegression #--------------- Binary Class ------ N = 300 np.random.seed(1) X = np.random.randn(N,2) y = np.random.randint(0,2,N) y.sort() X[y==0,:]+=2 # just creating classes a little far print(X.shape, y.shape) #(300, 2) (300,) model = LogisticRegression(alpha=0.1) model.fit(X,y,max_itr=1000) yp = model.predict(X) ypr = model.predict_proba(X) print('Accuracy : ',np.mean(yp==y)) print('Loss : ',model.Loss(y,ypr)) #Accuracy : 0.96 #Loss : 0.07046678918014998 fig = plt.figure(figsize=(12,5)) gs = fig.add_gridspec(2,3) ax1 = fig.add_subplot(gs[0, 0]) ax2 = fig.add_subplot(gs[1, 0]) ax3 = fig.add_subplot(gs[:, 1]) ax4 = fig.add_subplot(gs[:, 2]) model.plot_Lcurve(ax=ax1) model.plot_weights(ax=ax2) model.plot_weights2(ax=ax3) model.plot_boundries(X,y,alphaP=1,ax=ax4) plt.tight_layout() plt.show()
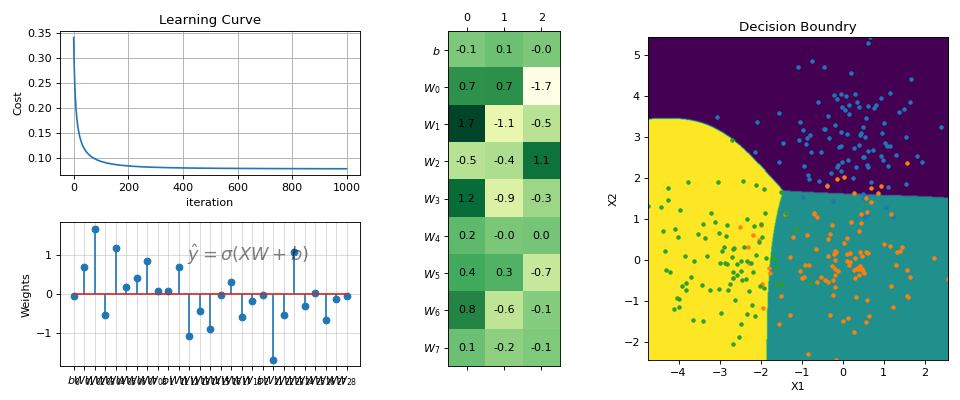
# ------- Multi Class ------ N =300 X = np.random.randn(N,2) y = np.random.randint(0,3,N) y.sort() X[y==0,1]+=3 X[y==2,0]-=3 print(X.shape, y.shape) #(300, 2) (300,) model = LogisticRegression(alpha=0.1,polyfit=True,degree=3,lambd=0,FeatureNormalize=True) model.fit(X,y,max_itr=1000) yp = model.predict(X) ypr = model.predict_proba(X) print('Accuracy : ',np.mean(yp==y)) print('Loss : ',model.Loss(model.oneHot(y),ypr)) #Accuracy : 0.8833333333333333 #Loss : 0.08365012491975303 fig = plt.figure(figsize=(12,5)) gs = fig.add_gridspec(2,3) ax1 = fig.add_subplot(gs[0, 0]) ax2 = fig.add_subplot(gs[1, 0]) ax3 = fig.add_subplot(gs[:, 1]) ax4 = fig.add_subplot(gs[:, 2]) model.plot_Lcurve(ax=ax1) model.plot_weights(ax=ax2) model.plot_weights2(ax=ax3) model.plot_boundries(X,y,alphaP=1,ax=ax4) plt.tight_layout() plt.show()
Methods
Loss(y, yp)Loss Function
Normalize(X)Normalization
PolyFeature(X[, degree])Polynomial Feature Extrations
fit(X, y[, max_itr, verbose, printAt, warm])Fitting the model
Get Weights
Get Weights
oneHot(y)Convert y to One hot vector
plot_Lcurve([ax])Plot Learning Curve
plot_boundries(X, y[, ax, density, ...])Plot Boundaries
plot_weights([ax, show_eq, fontsize, alpha])Plot Weights
plot_weights2([ax, fontsize, grid])Plot Weights as matrix
predict(X)Prediction
Predict probability
Regularization
sigmoid(z)Sigmoid function
softmax(z)Softmax function
- Loss(y, yp)¶
Loss Function
- Normalize(X)¶
Normalization
- PolyFeature(X, degree=2)¶
Polynomial Feature Extrations
- Parameters:
- X: 2d-array (n,nf)
n samples, nf features
- degree: int, default=2
degree of polynomials to comput
for degree of 2, f1**2, f2**2 … is computed and f1*f2*f3 ..
- Returns:
- Xp: 2d-array (n,mf)
new feature matrix with polynomial features
- fit(X, y, max_itr=-1, verbose=0, printAt=100, warm=True)¶
Fitting the model
- Parameters:
- Xndarray
(N,nf),
- yndarray of int (N,)
y \(\in\) [0,1,…]
- max_itr: int,
number of iteration, if -1, set to 10000
- verbose: int,0,
silent, 1, minimum verbosity, 11, debug mode
- warm: bool,
if false, weights will be reinitialize, else, last trained weights will be used
- getWeights()¶
Get Weights
- getWeightsAsList()¶
Get Weights
- Returns:
- bW: 2d-array
weights : [b, W]
- Wl: list of str
label of weights
- oneHot(y)¶
Convert y to One hot vector
- plot_Lcurve(ax=None)¶
Plot Learning Curve
- plot_boundries(X, y, ax=None, density=500, hardbound=False, alphaP=1, alphaB=1)¶
Plot Boundaries
Only for 2D dataset
- Parameters:
- X: features matirx
- y: labels
- ax: plt axis, default None
- density: pixel density for plot
- hardbound: bool, default=False
if True, then final classification decisions are used
else classification probalities are used
- plot_weights(ax=None, show_eq=True, fontsize=16, alpha=0.5)¶
Plot Weights
- Parameters:
- ax: default None,
axis to plot, if not provided, then fig, ax = plt.subplots()
- show_eq: bool, default=True
if true, equation is written on plot as text
- fontsize: fontsize of equation
- alpha: alpha for equation text
- plot_weights2(ax=None, fontsize=10, grid=True)¶
Plot Weights as matrix
- predict(X)¶
Prediction
- predict_proba(X)¶
Predict probability
- regularization(W)¶
Regularization
- sigmoid(z)¶
Sigmoid function
- softmax(z)¶
Softmax function