spkit.friedrichs_mollifier_kernel¶
- spkit.friedrichs_mollifier_kernel(window_size, s=1, p=2, r=0.999)¶
Mollifier: Kurt Otto Friedrichs
Mollifier: Kurt Otto Friedrichs
Generalized function
\[f(x) = exp(-s/(1-|x|**p)) for |x|<1, x \in [-r, r]\]Convolving with a mollifier, signals’s sharp features are smoothed, while still remaining close to the original nonsmooth (generalized) signals.
Intuitively, given a function which is rather irregular, by convolving it with a mollifier the function gets “mollified”.
This function is infinitely differentiable, non analytic with vanishing derivative for |x| = 1, can be therefore used as mollifier as described in [1]. This is a positive and symmetric mollifier.[15]
- Parameters:
- window_size: int, size of windows
- s: scaler, s>0, default=1,
Spread of the middle width, heigher the value of s, narrower the width
- p: scaler, p>0, default=2,
Order of flateness of the peak at the top,
p=2, smoother, p=1, triangulare type
Higher it is, more flat the peak.
- r: float, 0<r<1, default=0.999,
it is used to compute x = [-r, r]
recommonded to keep it r=0.999
- Returns:
- ker_mol: mollifier kernel
See also
gaussian_kernelGaussian Kernel
References
Examples
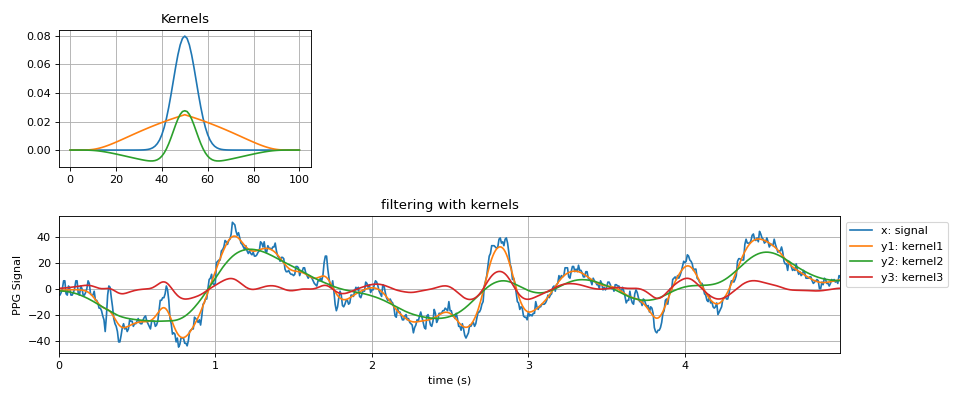
#sp.friedrichs_mollifier_kernel import numpy as np import matplotlib.pyplot as plt import spkit as sp x,fs = sp.data.ppg_sample(sample=1) x = x[:int(fs*5)] x = x - x.mean() t = np.arange(len(x))/fs kernel1 = sp.gaussian_kernel(window_length=101,sigma_scale=10) kernel2 = sp.friedrichs_mollifier_kernel(window_size=101,s=1,p=1) kernel3 = (kernel1 - kernel2)/2 y1 = sp.filter_with_kernel(x.copy(),kernel=kernel1) y2 = sp.filter_with_kernel(x.copy(),kernel=kernel2) y3 = sp.filter_with_kernel(x.copy(),kernel=kernel3) plt.figure(figsize=(12,5)) plt.subplot(212) plt.plot(t,x,label='x: signal') plt.plot(t,y1,label='y1: kernel1') plt.plot(t,y2,label='y2: kernel2') plt.plot(t,y3,label='y3: kernel3') plt.xlim([t[0],t[-1]]) plt.xlabel('time (s)') plt.ylabel('PPG Signal') plt.grid() plt.legend(bbox_to_anchor=(1,1)) plt.title('filtering with kernels') plt.subplot(231) plt.plot(kernel1,label='kernel1') plt.plot(kernel2,label='kernel2') plt.plot(kernel3,label='kernel3') plt.title('Kernels') plt.grid() plt.tight_layout() plt.show()