spkit.wavelet_filtering¶
- spkit.wavelet_filtering(x, wv='db3', threshold='optimal', filter_out_below=True, k=1.5, mode='elim', show=False, wpd_mode='symmetric', wpd_maxlevel=None, packetwise=False, WPD=False, lvl=[], verbose=False, fs=128.0, sf=1, IPR=[0.25, 0.75], figsize=(11, 6), plot_abs_coef=False)¶
Wavelet Filtering
Wavelet Filtering
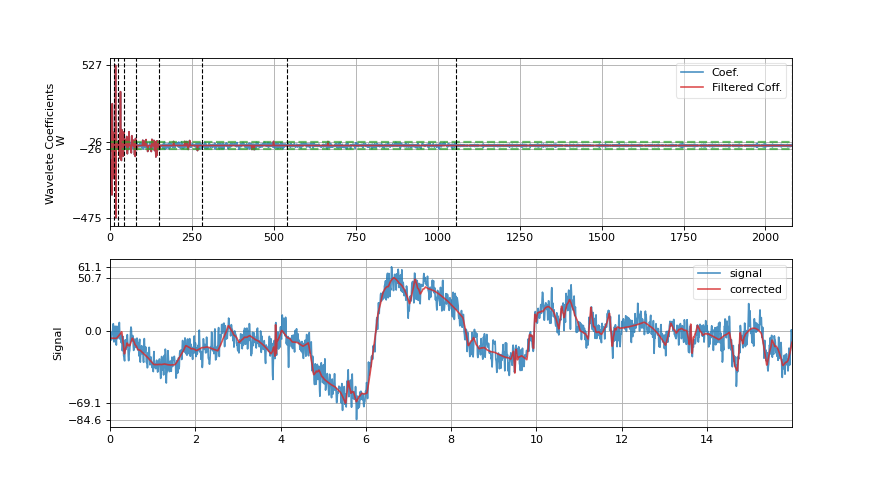
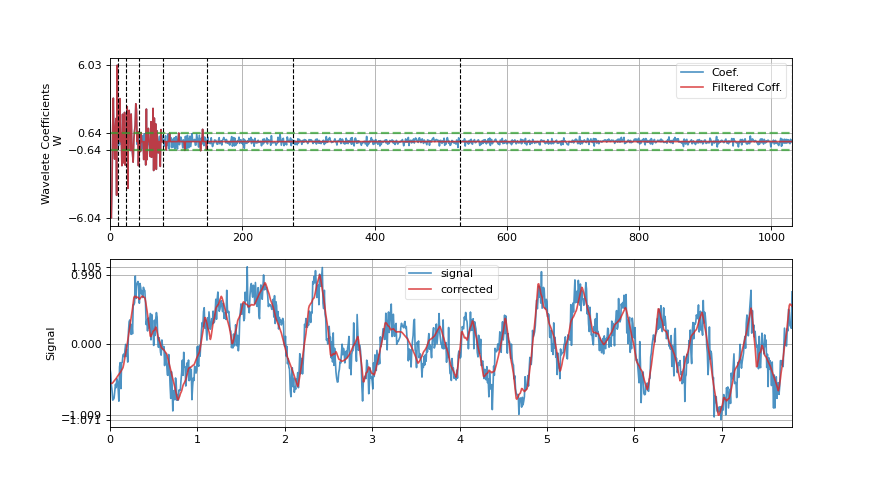
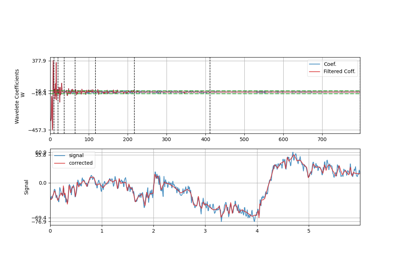
Wavelet filtering is applied on signal by decomposing signal into wavelet domain, filtering out wavelet coefficients and reconstructing signal back. Signal is decompose using DWT with wavelet function specified as wv (e.g. db3), and filtering out coefficients using by
threshold,mode, andfilter_out_belowarguments.- Wavelet Transform
\(W(k) = DWT(x(n))\)
Filtering (with
elimmode) - \(W(k)=0\)if \(|W(k)|<=threshold\) and if filter_out_below = True
if \(|W(k)|>threshold\) and if filter_out_below = False
- Reconstruction
\(x'(n) = IDWT(W(k))\)
- Parameters:
- x: 1d array
- Threshold Computation method:
- threshold: ‘str’ or float, default - ‘optimal’
if str, method to compute threshold, example : ‘optimal’, ‘sd’, ‘iqr’
‘optimal’: threshold = sig*sqrt(2logN), sig = median(|w|)/0.6745
‘sd’ : threshold = k*SD(w)
‘iqr’: threshold = q3+kr, threshold_l =q1-kr, where r = IQR(w) #Tukey’s fences
‘ttt’: Modified Thompson Tau test (ttt) # TODO
- wvstr, ‘db3’(default)
Wavelet family
{‘db3’…..’db38’, ‘sym2…..sym20’, ‘coif1…..coif17’, ‘bior1.1….bior6.8’, ‘rbio1.1…rbio6.8’, ‘dmey’}
- mode: str, default = ‘elim’
‘elim’ - remove the coeeficient (by zering out),
‘clip’ - cliping the coefficient to threshold
- filter_out_below: bool, default True,
if true, wavelet coefficient below threshold are eliminated else obove threshold
- wpd_mode = str, default ‘symmetric’
Wavelet Decomposition modes
[‘zero’, ‘constant’, ‘symmetric’, ‘periodic’, ‘smooth’, ‘periodization’]
- wpd_maxlevel: int
- level of decomposition,
if None, max level posible is used
- packetwise: bool, if true,
thresholding is applied to each packet/level individually, else globally
- WPD: if true,
WPD is applied as wavelet transform
- lvl: list
list of levels/packets apply the thresholding, if empty, applied to all the levels/packets
- show: bool, deafult=False,
if to plot figure, it True, following are used
figsize: default=(11,6), size of figure plot_abs_coef: bool, deafult=False,
if True,plot abs coefficient value, else signed
- Returns:
- xR: filtered signal, same size as x
See also
wavelet_filtering_winapplying wavelet filtering on smaller windows
filter_Xspectral filtering
Notes
#TODO
References
wikipedia -
Examples
import spkit as sp x,fs = sp.load_data.eeg_sample_1ch() xr = sp.wavelet_filtering(x,fs=fs, wv='db3', threshold='optimal',show=True) ################# import numpy as np import matplotlib.pyplot as plt import spkit as sp x = sp.create_signal_1d(n=1000,seed=1,sg_polyorder=5, sg_winlen=11) x = sp.add_noise(x,snr_db=10) xr = sp.wavelet_filtering(x.copy(),wv='db3',threshold='optimal',show=True)