spkit.geometry.get_adjacency_matrix_kNN¶
- spkit.geometry.get_adjacency_matrix_kNN(V, K=5, remove_self_con=True, verbose=False, ignore_matrix=False)¶
Create Adjacency Index Matrix based on Euclidean distance kNN
- Parameters:
- V: Points, (n,d)
n-points in d-dimentional space
- K: int,
number of nearest neibhbour
- remove_self_con: bool,
if true, self point is excluded from neareast neighbours
- Returns:
- AdjM: Adjacency Index Matrix,
(n, K) shape, index of neareast points, in accending order of distance
- node2C: dict,
Dictionary of node to connection list node2C[node_a] = list_of_nodes_connected_to_node_a
Examples
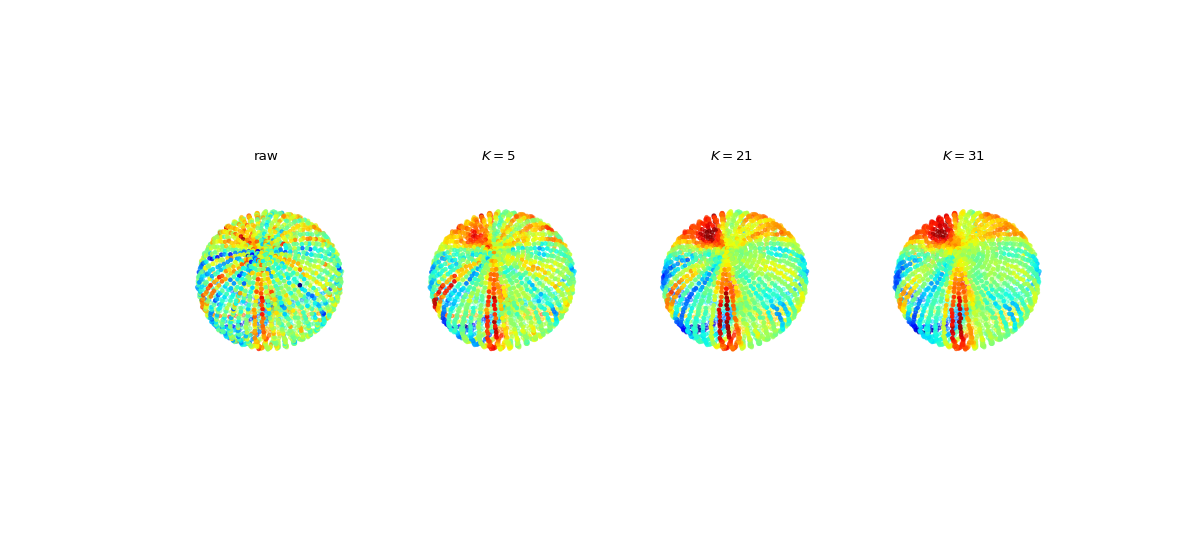
#sp.geometry.get_adjacency_matrix_kNN import numpy as np import matplotlib.pyplot as plt import spkit as sp V = sp.geometry.get_ellipsoid(n1=50, n2=50, rx=1, ry=2, rz=1,) V += 0.01*np.random.randn(V.shape[0],V.shape[1]) X = sp.create_signal_1d(V.shape[0],bipolar=False,sg_winlen=21,sg_polyorder=2,seed=1) X += 0.1*np.random.randn(X.shape[0]) AdjM1, _ = sp.geometry.get_adjacency_matrix_kNN(V, K=5) AdjM2, _ = sp.geometry.get_adjacency_matrix_kNN(V, K=21) AdjM3, _ = sp.geometry.get_adjacency_matrix_kNN(V, K=31) Y1 = sp.graph_filter_adj(X,AdjM1,ftype='mean',exclude_self=False) Y2 = sp.graph_filter_adj(X,AdjM2,ftype='mean',exclude_self=False) Y3 = sp.graph_filter_adj(X,AdjM3,ftype='mean',exclude_self=False) fig, ax = plt.subplots(1,4,subplot_kw={"projection": "3d"},figsize=(15,7)) Ys =[X, Y1, Y2, Y3] TITLEs = ['raw', r'$K=5$',r'$K=21$', r'$K=31$'] for i in range(4): ax[i].scatter3D(V[:,0], V[:,1], V[:,2], c=Ys[i], cmap='jet',s=10) ax[i].axis('off') ax[i].view_init(elev=60, azim=45, roll=15) ax[i].set_xlim([-1,1]) ax[i].set_ylim([-2,2]) ax[i].set_zlim([-1,1]) ax[i].set_title(TITLEs[i]) plt.subplots_adjust(hspace=0,wspace=0) plt.show()