spkit.eeg.topomap¶
- spkit.eeg.topomap(data, pos=None, ch_names=None, res=64, Zi=None, show=True, return_im=False, standard='1020', style='spkit', **kwargs)¶
Display Topographical Map of EEG, with given values
- Parameters:
- data: 1d-array
1d-array of values for the each electrode.
- pos: 2d-array, deafult=None
2D projection points for each electroed
- computed using one of either
s1020_get_epos2d,s1010_get_epos2d,s1005_get_epos2d or provided by custom system
- computed using one of either
MUST BE same number of points as data points, e.g. pos.shape[0]==data.shape[0]
IF
None, then computed usingch_namessystemstyleprovided.
- ch_names: list of str, default=None
name of the channels/electrodes
should be same size as data.shape[0]
IF
posis None, this is used to compute projection points (pos), in that case, make sure to provide valid channel names and respective system. or compute usings1020_get_epos2d,s1010_get_epos2d,s1005_get_epos2dThis list is also used to shownames of sensors, if
shownamesis True.
Note
IMPORTANT Either
posor validch_namesshould be provided- res: int, default=64
resolution of image to be generated, (res,res)
- Zi: 2d-array, default None,
Pre-computed Image of topographic map.
if provided, then
datais ignored, else image is generated using data-points
- show: bool, defult=True
if False, then plot is not produced
useful, when interested in only generating Zi images
- return_im: bool, default=False
if True, in addtion to Zi (generate Image),
imobject is also returned
- standard: str, default =’1020’
it is used ONLY if
posis Nonestandard to extract pos from ch_names
- style: str, default = ‘spkit’
it is used ONLY if
posis None and standard is ‘1020’style to extract pos from ch_names
check
s1020_get_epos2d
- kwargs:
There are other arguments which can be supplied to modify the topomap disply
- warn: bool, default True
set it to False, to supress the warnings of change names
- c:scalar,default=0.5
shifting origin, only used if
shift_originis True
- s:scalar,default=0.85
scale the electrod postions by the factor
s, higher it is more spreaded they are
- (fx, fy):scalar,default=(0.5, 0.5)
x-lim, y-lim of grid,
- (rx,ry):scalar,default=(1,1)
radius of the ellips to clip, higher it is more outter area is covered
- shift_origin=False
If True, center point of electrode poisition will be the origin = (0,0)
it uses
cparameter to shift the originpos -= c*([dx, dy])
if
c=1, new origin will be at 0,0
axes=None: Axes to use for plotting, if None, then created using plt.gca()
- For Image display, plt.imshow kwargs:
(cmap=’jet’,origin=’lower’,aspect=’equal’,vmin=None,vmax=None,interpolation=None)
- contours=True: contour lines
if True,
contours_ncontours are shown with linewidth ofcontours_lw
- showsensors=True: sensor location
display sensors/eletrodes as dots
add cicles of sensors using
sensor_prop
- shownames=True
if True, show them, using
fontdictandfont_propdefault fontdict=None, font_prop=dict()
- showhead=True
If True, show head, using
head_prop(default head_prop =dict(markersize=30))it is passed to ax.plot as kwargs
- showbound=True: show boundary
uses bound_prop=dict(rx=0.85,ry=0.85,xy=(0,0),color=’k’,alpha=0.5)
could be used to show oval shape head, with rx,ry = 0.85,0.9
- show_vhlines=True: lines
show verticle and horizontal lines passing through origin
- show_colorbar=False, colorbar
if True, show colorbar with label as
colorbar_label
- match_shed=True: boundary
if True, center of boundary is same as center of grid.
- Returns:
- Zi: 2d-array
2D full Grid as image, without circular crop.
Obatained from Inter/exterpolation
- im: image object
returned from im = ax.imshow()
only of
return_imis True
See also
Gen_SSFI
Examples
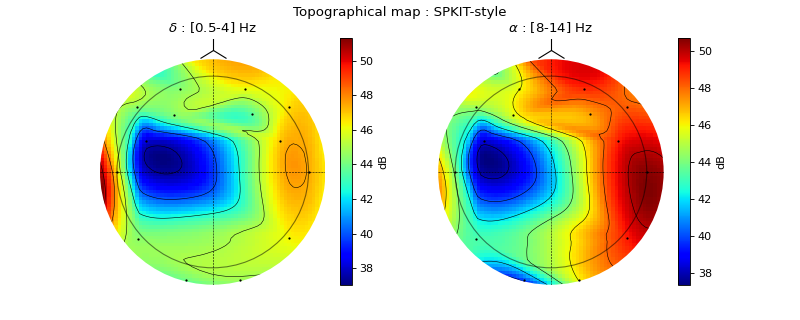
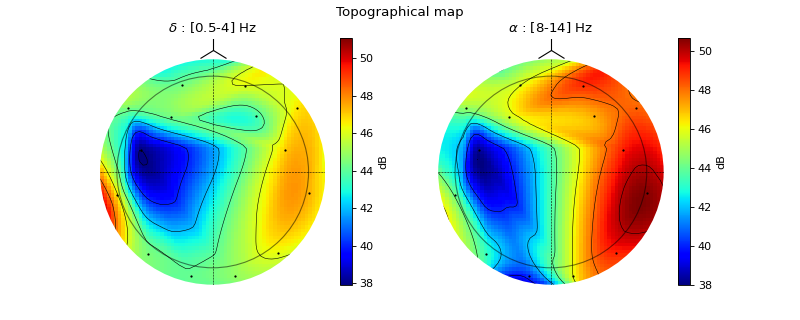
#sp.eeg.topomap import numpy as np import matplotlib.pyplot as plt import spkit as sp X,fs, ch_names = sp.data.eeg_sample_14ch() X = sp.filterDC_sGolay(X, window_length=fs//3+1) Px,Pm,Pd = sp.eeg.rhythmic_powers(X=X,fs=fs,fBands=[[4],[8,14]],Sum=True,Mean=True,SD =True) Px = 20*np.log10(Px) pos1, ch = sp.eeg.s1020_get_epos2d(ch_names,style='eeglab-mne') pos2, ch = sp.eeg.s1020_get_epos2d(ch_names,style='spkit') fig, ax = plt.subplots(1,2,figsize=(10,4)) Z1i,im1 = sp.eeg.topomap(pos=pos1,data=Px[0],axes=ax[0],return_im=True) Z2i,im2 = sp.eeg.topomap(pos=pos1,data=Px[1],axes=ax[1],return_im=True) ax[0].set_title(r'$\delta$ : [0.5-4] Hz') ax[1].set_title(r'$\alpha$ : [8-14] Hz') plt.colorbar(im1, ax=ax[0],label='dB') plt.colorbar(im2, ax=ax[1],label='dB') plt.suptitle('Topographical map') plt.show()
fig, ax = plt.subplots(1,2,figsize=(10,4)) Z1i,im1 = sp.eeg.topomap(pos=pos2,data=Px[0],axes=ax[0],return_im=True) Z2i,im2 = sp.eeg.topomap(pos=pos2,data=Px[1],axes=ax[1],return_im=True) ax[0].set_title(r'$\delta$ : [0.5-4] Hz') ax[1].set_title(r'$\alpha$ : [8-14] Hz') plt.colorbar(im1, ax=ax[0],label='dB') plt.colorbar(im2, ax=ax[1],label='dB') plt.suptitle('Topographical map : SPKIT-style') plt.show()