spkit.eeg.gen_ssfi¶
- spkit.eeg.gen_ssfi(PX, pos, res=64, NormalizeEachBand=False, verbose=1)¶
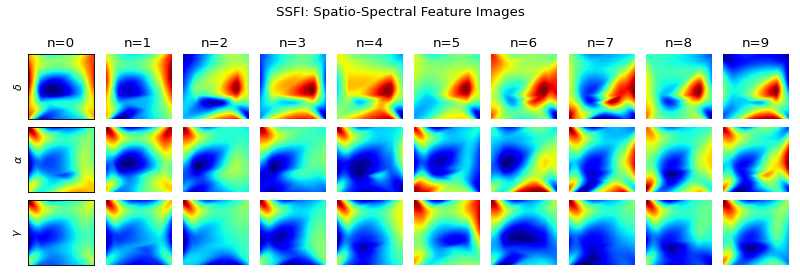
Generating Spatio-Spectral Feature Image (SSFI)
Spatio-Spectral Feature Images
Input - PX : (100,6,14) or (100,6*14)
Output - Ximg : (100,6,64,64)
- Parameters:
- PX: 2D-array or 3D-array,
Power values of the each band and each channel
- if 2D-array
shape of (nt , N*M)
where nt=number of segments, N=number of frequency bands, M=number of channels
- e.g:
for 5 chennels, three bands, power values of each band is in sequence - PX[0] = [1,2,3,4,5, 1,2,3,4,5, 6,1,2,3,4,5] - PX.shape = (100, 3*5), 100 time instant, 3 bands and 5 channels
- if 3D-array
shape of (nt, N, M)
where nt=number of segments, N=number of frequency bands, M=number of channels
- e.g:
for 5 chennels, three bands, power values of each band is in sequence - PX.shape = (100, 3, 14), 100 time instant, 3 bands and 14 channels
- pos: 2D projections
shape = (M,2)
number of channel M same in order as values
- res: int default=64
resolution of SSFI
- NormalizeEachBand: bool, Default=False
If True, power values of each band are normalised
it is useful, if distribution if more improtant than power values
- Returns:
- Ximg: 3D-Array
(nt,N, res, res)
References
[1] Bajaj N, Requena Carrión J., (2023, August). Deep Representation of EEG Signals Using Spatio-Spectral Feature Images. Applied Sciences. 2023; 13(17):9825. https://doi.org/10.3390/app13179825. [Link]
Examples
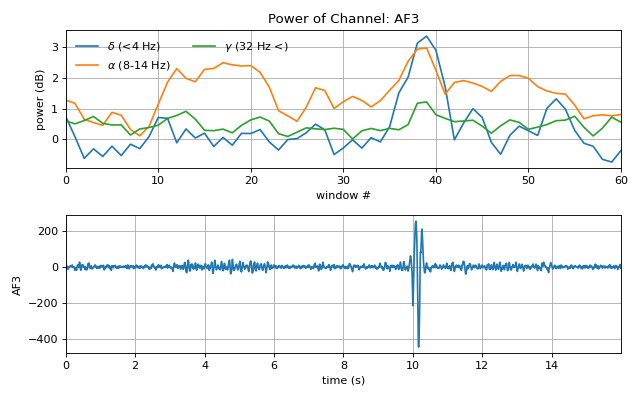
#sp.eeg.gen_ssfi import numpy as np import matplotlib.pyplot as plt import spkit as sp X,fs, ch_names = sp.data.eeg_sample_14ch() X = sp.filterDC_sGolay(X, window_length=fs//3+1) t = np.arange(X.shape[0])/fs Pxt, _, _ = sp.eeg.rhythmic_powers_win(X, winsize=128,overlap=32,fBands=[[4],[8,14],[32,47]],Sum=True) Pxt = np.log10(Pxt) pos, ch = sp.eeg.s1020_get_epos2d(ch_names,style='spkit') Ximg = sp.eeg.gen_ssfi(Pxt,pos=pos, res=64) plt.figure(figsize=(8,5)) plt.subplot(211) plt.plot(Pxt[:,:,0], label=[r'$\delta$ (<4 Hz)',r'$\alpha$ (8-14 Hz)',r'$\gamma$ (32 Hz <)',]) plt.xlabel('window #') plt.ylabel('power (dB)') plt.title(f'Power of Channel: {ch_names[0]}') plt.xlim([0,Pxt.shape[0]-1]) plt.grid() plt.legend(ncol=2,frameon=False) plt.subplot(212) plt.plot(t,X[:,0]) plt.grid() plt.xlim([t[0],t[-1]]) plt.xlabel('time (s)') plt.ylabel(ch_names[0]) plt.tight_layout() plt.show()
plt.figure(figsize=(10,3.5)) for i in range(10): plt.subplot(3,10,i+1) plt.imshow(Ximg[i,0],cmap='jet') plt.title(f'n={i}') if i==0: plt.ylabel(r'$\delta$') plt.xticks([]) plt.yticks([]) else: plt.axis('off') plt.subplot(3,10,10+i+1) plt.imshow(Ximg[i,1],cmap='jet') if i==0: plt.ylabel(r'$\alpha$') plt.xticks([]) plt.yticks([]) else: plt.axis('off') plt.subplot(3,10,20+i+1) plt.imshow(Ximg[i,2],cmap='jet') if i==0: plt.ylabel(r'$\gamma$') plt.xticks([]) plt.yticks([]) else: plt.axis('off') plt.suptitle('SSFI: Spatio-Spectral Feature Images') plt.tight_layout() plt.show()